TRF2 rescues telomere attrition and prolongs cell survival in Duchenne muscular dystrophy cardiomyocytes derived from human iPSCs
- PMID: 36719921
- PMCID: PMC9963063
- DOI: 10.1073/pnas.2209967120
TRF2 rescues telomere attrition and prolongs cell survival in Duchenne muscular dystrophy cardiomyocytes derived from human iPSCs
Abstract
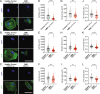
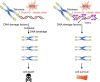
Duchenne muscular dystrophy (DMD) is a severe muscle wasting disease caused by the lack of dystrophin. Heart failure, driven by cardiomyocyte death, fibrosis, and the development of dilated cardiomyopathy, is the leading cause of death in DMD patients. Current treatments decrease the mechanical load on the heart but do not address the root cause of dilated cardiomyopathy: cardiomyocyte death. Previously, we showed that telomere shortening is a hallmark of DMD cardiomyocytes. Here, we test whether prevention of telomere attrition is possible in cardiomyocytes differentiated from patient-derived induced pluripotent stem cells (iPSC-CMs) and if preventing telomere shortening impacts cardiomyocyte function. We observe reduced cell size, nuclear size, and sarcomere density in DMD iPSC-CMs compared with healthy isogenic controls. We find that expression of just one telomere-binding protein, telomeric repeat-binding factor 2 (TRF2), a core component of the shelterin complex, prevents telomere attrition and rescues deficiencies in cell size as well as sarcomere density. We employ a bioengineered platform to micropattern cardiomyocytes for calcium imaging and perform Southern blots of telomere restriction fragments, the gold standard for telomere length assessments. Importantly, preservation of telomere lengths in DMD cardiomyocytes improves their viability. These data provide evidence that preventing telomere attrition ameliorates deficits in cell morphology, activation of the DNA damage response, and premature cell death, suggesting that TRF2 is a key player in DMD-associated cardiac failure.
Keywords: Duchenne muscular dystrophy; cardiomyocytes; induced pluripotent stem cells; telomere.
Conflict of interest statement
The authors declare no competing interest.
Figures




Similar articles
-
Modeling and study of the mechanism of dilated cardiomyopathy using induced pluripotent stem cells derived from individuals with Duchenne muscular dystrophy.Dis Model Mech. 2015 May;8(5):457-66. doi: 10.1242/dmm.019505. Epub 2015 Mar 19. Dis Model Mech. 2015. PMID: 25791035 Free PMC article.
-
Electrophysiological abnormalities in induced pluripotent stem cell-derived cardiomyocytes generated from Duchenne muscular dystrophy patients.J Cell Mol Med. 2019 Mar;23(3):2125-2135. doi: 10.1111/jcmm.14124. Epub 2019 Jan 8. J Cell Mol Med. 2019. PMID: 30618214 Free PMC article.
-
Absence of full-length dystrophin impairs normal maturation and contraction of cardiomyocytes derived from human-induced pluripotent stem cells.Cardiovasc Res. 2020 Feb 1;116(2):368-382. doi: 10.1093/cvr/cvz109. Cardiovasc Res. 2020. PMID: 31049579 Free PMC article.
-
Modeling Duchenne Muscular Dystrophy Cardiomyopathy with Patients' Induced Pluripotent Stem-Cell-Derived Cardiomyocytes.Int J Mol Sci. 2023 May 12;24(10):8657. doi: 10.3390/ijms24108657. Int J Mol Sci. 2023. PMID: 37240001 Free PMC article. Review.
-
Cardiac phenotype of Duchenne Muscular Dystrophy: insights from cellular studies.J Mol Cell Cardiol. 2013 May;58:217-24. doi: 10.1016/j.yjmcc.2012.12.009. Epub 2012 Dec 20. J Mol Cell Cardiol. 2013. PMID: 23261966 Free PMC article. Review.
Cited by
-
Clinically Actionable Topical Strategies for Addressing the Hallmarks of Skin Aging: A Primer for Aesthetic Medicine Practitioners.Cureus. 2024 Jan 19;16(1):e52548. doi: 10.7759/cureus.52548. eCollection 2024 Jan. Cureus. 2024. PMID: 38371024 Free PMC article. Review.
-
Induced Pluripotent (iPSC) and Mesenchymal (MSC) Stem Cells for In Vitro Disease Modeling and Regenerative Medicine.Int J Mol Sci. 2025 Jun 11;26(12):5617. doi: 10.3390/ijms26125617. Int J Mol Sci. 2025. PMID: 40565081 Free PMC article. Review.
-
Roles of chromatin and genome instability in cellular senescence and their relevance to ageing and related diseases.Nat Rev Mol Cell Biol. 2024 Dec;25(12):979-1000. doi: 10.1038/s41580-024-00775-3. Epub 2024 Oct 3. Nat Rev Mol Cell Biol. 2024. PMID: 39363000 Review.
-
Comprehensive review on gene mutations contributing to dilated cardiomyopathy.Front Cardiovasc Med. 2023 Dec 1;10:1296389. doi: 10.3389/fcvm.2023.1296389. eCollection 2023. Front Cardiovasc Med. 2023. PMID: 38107262 Free PMC article. Review.
-
Ryanodine receptor dysfunction causes senescence and fibrosis in Duchenne dilated cardiomyopathy.J Cachexia Sarcopenia Muscle. 2024 Apr;15(2):536-551. doi: 10.1002/jcsm.13411. Epub 2024 Jan 14. J Cachexia Sarcopenia Muscle. 2024. PMID: 38221511 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

