The ribose methylation enzyme FTSJ1 has a conserved role in neuron morphology and learning performance
- PMID: 36720500
- PMCID: PMC9889914
- DOI: 10.26508/lsa.202201877
The ribose methylation enzyme FTSJ1 has a conserved role in neuron morphology and learning performance
Abstract
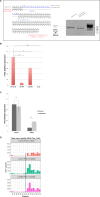
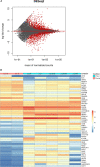
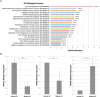
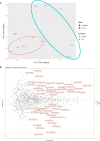
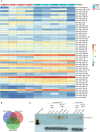
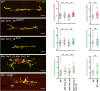
FTSJ1 is a conserved human 2'-O-methyltransferase (Nm-MTase) that modifies several tRNAs at position 32 and the wobble position 34 in the anticodon loop. Its loss of function has been linked to X-linked intellectual disability (XLID), and more recently to cancers. However, the molecular mechanisms underlying these pathologies are currently unclear. Here, we report a novel FTSJ1 pathogenic variant from an X-linked intellectual disability patient. Using blood cells derived from this patient and other affected individuals carrying FTSJ1 mutations, we performed an unbiased and comprehensive RiboMethSeq analysis to map the ribose methylation on all human tRNAs and identify novel targets. In addition, we performed a transcriptome analysis in these cells and found that several genes previously associated with intellectual disability and cancers were deregulated. We also found changes in the miRNA population that suggest potential cross-regulation of some miRNAs with these key mRNA targets. Finally, we show that differentiation of FTSJ1-depleted human neural progenitor cells into neurons displays long and thin spine neurites compared with control cells. These defects are also observed in Drosophila and are associated with long-term memory deficits. Altogether, our study adds insight into FTSJ1 pathologies in humans and flies by the identification of novel FTSJ1 targets and the defect in neuron morphology.
© 2023 Brazane et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




References
-
- Angelova MT, Dimitrova DG, Da Silva B, Marchand V, Jacquier C, Achour C, Brazane M, Goyenvalle C, Bourguignon-Igel V, Shehzada S, et al. (2020) tRNA 2’-O-methylation by a duo of TRM7/FTSJ1 proteins modulates small RNA silencing in Drosophila. Nucleic Acids Res 48: 2050–2072. 10.1093/nar/gkaa002 - DOI - PMC - PubMed
-
- Bartoli KM, Schaening C, Carlile TM, Gilbert WV (2018) Conserved methyltransferase Spb1 targets mRNAs for regulated modification with 2′-O-methyl ribose. BioRxiv. 10.1101/271916 (Preprint posted March 8, 2018). - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
