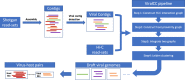
ViralCC retrieves complete viral genomes and virus-host pairs from metagenomic Hi-C data
- PMID: 36720887
- PMCID: PMC9889337
- DOI: 10.1038/s41467-023-35945-y
ViralCC retrieves complete viral genomes and virus-host pairs from metagenomic Hi-C data
Abstract
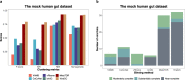
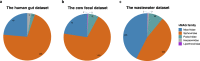
The introduction of high-throughput chromosome conformation capture (Hi-C) into metagenomics enables reconstructing high-quality metagenome-assembled genomes (MAGs) from microbial communities. Despite recent advances in recovering eukaryotic, bacterial, and archaeal genomes using Hi-C contact maps, few of Hi-C-based methods are designed to retrieve viral genomes. Here we introduce ViralCC, a publicly available tool to recover complete viral genomes and detect virus-host pairs using Hi-C data. Compared to other Hi-C-based methods, ViralCC leverages the virus-host proximity structure as a complementary information source for the Hi-C interactions. Using mock and real metagenomic Hi-C datasets from several different microbial ecosystems, including the human gut, cow fecal, and wastewater, we demonstrate that ViralCC outperforms existing Hi-C-based binning methods as well as state-of-the-art tools specifically dedicated to metagenomic viral binning. ViralCC can also reveal the taxonomic structure of viruses and virus-host pairs in microbial communities. When applied to a real wastewater metagenomic Hi-C dataset, ViralCC constructs a phage-host network, which is further validated using CRISPR spacer analyses. ViralCC is an open-source pipeline available at https://github.com/dyxstat/ViralCC .
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
HiCBin: binning metagenomic contigs and recovering metagenome-assembled genomes using Hi-C contact maps.Genome Biol. 2022 Feb 28;23(1):63. doi: 10.1186/s13059-022-02626-w. Genome Biol. 2022. PMID: 35227283 Free PMC article.
-
Novel canine high-quality metagenome-assembled genomes, prophages and host-associated plasmids provided by long-read metagenomics together with Hi-C proximity ligation.Microb Genom. 2022 Mar;8(3):000802. doi: 10.1099/mgen.0.000802. Microb Genom. 2022. PMID: 35298370 Free PMC article.
-
HiFine: integrating Hi-C-based and shotgun-based methods to refine binning of metagenomic contigs.Bioinformatics. 2022 May 26;38(11):2973-2979. doi: 10.1093/bioinformatics/btac295. Bioinformatics. 2022. PMID: 35482530 Free PMC article.
-
Recovering metagenome-assembled genomes from shotgun metagenomic sequencing data: Methods, applications, challenges, and opportunities.Microbiol Res. 2022 Jul;260:127023. doi: 10.1016/j.micres.2022.127023. Epub 2022 Apr 8. Microbiol Res. 2022. PMID: 35430490 Review.
-
Computational Tools for the Analysis of Uncultivated Phage Genomes.Microbiol Mol Biol Rev. 2022 Jun 15;86(2):e0000421. doi: 10.1128/mmbr.00004-21. Epub 2022 Mar 21. Microbiol Mol Biol Rev. 2022. PMID: 35311574 Free PMC article. Review.
Cited by
-
Solving genomic puzzles: computational methods for metagenomic binning.Brief Bioinform. 2024 Jul 25;25(5):bbae372. doi: 10.1093/bib/bbae372. Brief Bioinform. 2024. PMID: 39082646 Free PMC article. Review.
-
VirGrapher: a graph-based viral identifier for long sequences from metagenomes.Brief Bioinform. 2024 Jan 22;25(2):bbae036. doi: 10.1093/bib/bbae036. Brief Bioinform. 2024. PMID: 38343326 Free PMC article.
-
Gut phages and their interactions with bacterial and mammalian hosts.J Bacteriol. 2025 Feb 20;207(2):e0042824. doi: 10.1128/jb.00428-24. Epub 2025 Jan 23. J Bacteriol. 2025. PMID: 39846747 Free PMC article. Review.
-
MetaHiCNet: a web server for normalizing and visualizing microbial Hi-C interaction networks.Nucleic Acids Res. 2025 Jul 7;53(W1):W383-W389. doi: 10.1093/nar/gkaf340. Nucleic Acids Res. 2025. PMID: 40287822 Free PMC article.
-
High-throughput single-cell sequencing of activated sludge microbiome.Environ Sci Ecotechnol. 2024 Sep 12;23:100493. doi: 10.1016/j.ese.2024.100493. eCollection 2025 Jan. Environ Sci Ecotechnol. 2024. PMID: 39430728 Free PMC article.
References
-
- Breitbart M, Rohwer F. Here a virus, there a virus, everywhere the same virus? Trends Microbiol. 2005;13:278–284. - PubMed
-
- Gobler CJ, Hutchins DA, Fisher NS, Cosper EM, Saňudo-Wilhelmy SA. Release and bioavailability of C, N, P Se, and Fe following viral lysis of a marine chrysophyte. Limnol. Oceanogr. 1997;42:1492–1504.
-
- Suttle CA. Marine viruses-major players in the global ecosystem. Nat. Rev. Microbiol. 2007;5:801–812. - PubMed
-
- Fuhrman JA. Marine viruses and their biogeochemical and ecological effects. Nature. 1999;399:541–548. - PubMed
-
- Jiao N, et al. Microbial production of recalcitrant dissolved organic matter: long-term carbon storage in the global ocean. Nat. Rev. Microbiol. 2010;8:593–599. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

