Integrated multiomics analysis to infer COVID-19 biological insights
- PMID: 36720931
- PMCID: PMC9888750
- DOI: 10.1038/s41598-023-28816-5
Integrated multiomics analysis to infer COVID-19 biological insights
Abstract
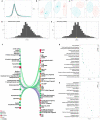
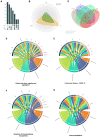
Three years after the pandemic, we still have an imprecise comprehension of the pathogen landscape and we are left with an urgent need for early detection methods and effective therapy for severe COVID-19 patients. The implications of infection go beyond pulmonary damage since the virus hijacks the host's cellular machinery and consumes its resources. Here, we profiled the plasma proteome and metabolome of a cohort of 57 control and severe COVID-19 cases using high-resolution mass spectrometry. We analyzed their proteome and metabolome profiles with multiple depths and methodologies as conventional single omics analysis and other multi-omics integrative methods to obtain the most comprehensive method that portrays an in-depth molecular landscape of the disease. Our findings revealed that integrating the knowledge-based and statistical-based techniques (knowledge-statistical network) outperformed other methods not only on the pathway detection level but even on the number of features detected within pathways. The versatile usage of this approach could provide us with a better understanding of the molecular mechanisms behind any biological system and provide multi-dimensional therapeutic solutions by simultaneously targeting more than one pathogenic factor.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- WHO. Coronavirus Disease (COVID-19) Dashboard., https://covid19.who.int/ (2021).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

