Jewel Beetle Opsin Duplication and Divergence Is the Mechanism for Diverse Spectral Sensitivities
- PMID: 36721951
- PMCID: PMC9937044
- DOI: 10.1093/molbev/msad023
Jewel Beetle Opsin Duplication and Divergence Is the Mechanism for Diverse Spectral Sensitivities
Abstract
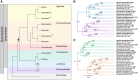
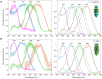
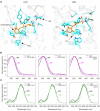
The evolutionary history of visual genes in Coleoptera differs from other well-studied insect orders, such as Lepidoptera and Diptera, as beetles have lost the widely conserved short-wavelength (SW) insect opsin gene that typically underpins sensitivity to blue light (∼440 nm). Duplications of the ancestral ultraviolet (UV) and long-wavelength (LW) opsins have occurred in many beetle lineages and have been proposed as an evolutionary route for expanded spectral sensitivity. The jewel beetles (Buprestidae) are a highly ecologically diverse and colorful family of beetles that use color cues for mate and host detection. In addition, there is evidence that buprestids have complex spectral sensitivity with up to five photoreceptor classes. Previous work suggested that opsin duplication and subfunctionalization of the two ancestral buprestid opsins, UV and LW, has expanded sensitivity to different regions of the light spectrum, but this has not yet been tested. We show that both duplications are likely unique to Buprestidae or the wider superfamily of Buprestoidea. To directly test photopigment sensitivity, we expressed buprestid opsins from two Chrysochroa species in Drosophila melanogaster and functionally characterized each photopigment type as UV- (356-357 nm), blue- (431-442 nm), green- (507-509 nm), and orange-sensitive (572-584 nm). As these novel opsin duplicates result in significantly shifted spectral sensitivities from the ancestral copies, we explored spectral tuning at four candidate sites using site-directed mutagenesis. This is the first study to directly test opsin spectral tuning mechanisms in the diverse and specious beetles.
Keywords: Drosophila; Buprestidae; Coleoptera; Insect vision; Spectral tuning; Visual pigment.
© The Author(s) 2023. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures

References
-
- Asenjo AB, Rim J, Oprian DD. 1994. Molecular determinants of human red/green color discrimination. Neuron. 12:1131–1138. - PubMed
-
- Briscoe AD, Chittka L. 2001. Evolution of colour vision in insects. Annu Rev Entomol. 46:471–510. - PubMed
-
- Chan T, Lee M, Sakmar TP. 1992. Introduction of hydroxyl-bearing amino acids causes bathochromic spectral shifts in rhodopsin. Amino acid substitutions responsible for red-green color pigment spectral tuning. J Biol Chem. 267:9478–9480. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

