Circulating Tumor DNA Is Prognostic in Intermediate-Risk Rhabdomyosarcoma: A Report From the Children's Oncology Group
- PMID: 36724417
- PMCID: PMC10150913
- DOI: 10.1200/JCO.22.00409
Circulating Tumor DNA Is Prognostic in Intermediate-Risk Rhabdomyosarcoma: A Report From the Children's Oncology Group
Abstract
Purpose: Novel biomarkers are needed to differentiate outcomes in intermediate-risk rhabdomyosarcoma (IR RMS). We sought to evaluate strategies for identifying circulating tumor DNA (ctDNA) in IR RMS and to determine whether ctDNA detection before therapy is associated with outcome.
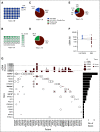
Patients and methods: Pretreatment serum and tumor samples were available from 124 patients with newly diagnosed IR RMS from the Children's Oncology Group biorepository, including 75 patients with fusion-negative rhabdomyosarcoma (FN-RMS) and 49 with fusion-positive rhabdomyosarcoma (FP-RMS) disease. We used ultralow passage whole-genome sequencing to detect copy number alterations and a new custom sequencing assay, Rhabdo-Seq, to detect rearrangements and single-nucleotide variants.
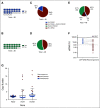
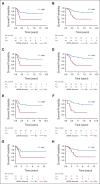
Results: We found that ultralow passage whole-genome sequencing was a method applicable to ctDNA detection in all patients with FN-RMS and that ctDNA was detectable in 13 of 75 serum samples (17%). However, the use of Rhabdo-Seq in FN-RMS samples also identified single-nucleotide variants, such as MYOD1L122R, previously associated with prognosis. Identification of pathognomonic translocations between PAX3 or PAX7 and FOXO1 by Rhabdo-Seq was the best method for measuring ctDNA in FP-RMS and detected ctDNA in 27 of 49 cases (55%). Patients with FN-RMS with detectable ctDNA at diagnosis had significantly worse outcomes than patients without detectable ctDNA (event-free survival, 33.3% v 68.9%; P = .0028; overall survival, 33.3% v 83.2%; P < .0001) as did patients with FP-RMS (event-free survival, 37% v 70%; P = .045; overall survival, 39.2% v 75%; P = .023). In multivariable analysis, ctDNA was independently associated with worse prognosis in FN-RMS but not in the smaller FP-RMS cohort.
Conclusion: Our study demonstrates that baseline ctDNA detection is feasible and is prognostic in IR RMS.
Conflict of interest statement
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (
Figures
Comment in
-
Detection of pretreatment circulating tumor DNA as a biomarker of poor outcome in intermediate-risk rhabdomyosarcoma.Transl Pediatr. 2024 May 31;13(5):869-872. doi: 10.21037/tp-24-7. Epub 2024 May 28. Transl Pediatr. 2024. PMID: 38840688 Free PMC article. No abstract available.
References
-
- US Department of Health and Human Services; National Cancer Institute : Cancer Incidence and Survival Among Children and Adolescents: United States SEER Program 1975-1995. 1999. http://www.crossref.org/deleted_DOI.html
-
- Arndt CAS, Stoner JA, Hawkins DS, et al. : Vincristine, actinomycin, and cyclophosphamide compared with vincristine, actinomycin, and cyclophosphamide alternating with vincristine, topotecan, and cyclophosphamide for intermediate-risk rhabdomyosarcoma: Children's Oncology Group Study D9803. J Clin Oncol 27:5182-5188, 2009 - PMC - PubMed
-
- Cescon DW, Bratman SV, Chan SM, et al. : Circulating tumor DNA and liquid biopsy in oncology. Nat Cancer 1:276-290, 2020 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

