Single-cell spatial immune landscapes of primary and metastatic brain tumours
- PMID: 36725935
- PMCID: PMC9931580
- DOI: 10.1038/s41586-022-05680-3
Single-cell spatial immune landscapes of primary and metastatic brain tumours
Abstract
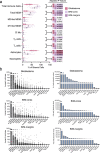
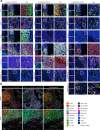
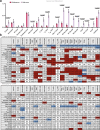
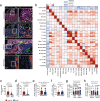
Single-cell technologies have enabled the characterization of the tumour microenvironment at unprecedented depth and have revealed vast cellular diversity among tumour cells and their niche. Anti-tumour immunity relies on cell-cell relationships within the tumour microenvironment1,2, yet many single-cell studies lack spatial context and rely on dissociated tissues3. Here we applied imaging mass cytometry to characterize the immunological landscape of 139 high-grade glioma and 46 brain metastasis tumours from patients. Single-cell analysis of more than 1.1 million cells across 389 high-dimensional histopathology images enabled the spatial resolution of immune lineages and activation states, revealing differences in immune landscapes between primary tumours and brain metastases from diverse solid cancers. These analyses revealed cellular neighbourhoods associated with survival in patients with glioblastoma, which we leveraged to identify a unique population of myeloperoxidase (MPO)-positive macrophages associated with long-term survival. Our findings provide insight into the biology of primary and metastatic brain tumours, reinforcing the value of integrating spatial resolution to single-cell datasets to dissect the microenvironmental contexture of cancer.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures








Comment in
-
Spatial proteomics of tumor microenvironments reveal why location matters.Nat Immunol. 2023 Apr;24(4):565-566. doi: 10.1038/s41590-023-01471-8. Nat Immunol. 2023. PMID: 36959293 No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

