Lost genome segments associate with trait diversity during rice domestication
- PMID: 36726089
- PMCID: PMC9893545
- DOI: 10.1186/s12915-023-01512-6
Lost genome segments associate with trait diversity during rice domestication
Abstract
Background: DNA mutations of diverse types provide the raw material required for phenotypic variation and evolution. In the case of crop species, previous research aimed to elucidate the changing patterns of repetitive sequences, single-nucleotide polymorphisms (SNPs), and small InDels during domestication to explain morphological evolution and adaptation to different environments. Additionally, structural variations (SVs) encompassing larger stretches of DNA are more likely to alter gene expression levels leading to phenotypic variation affecting plant phenotypes and stress resistance. Previous studies on SVs in rice were hampered by reliance on short-read sequencing limiting the quantity and quality of SV identification, while SV data are currently only available for cultivated rice, with wild rice largely uncharacterized. Here, we generated two genome assemblies for O. rufipogon using long-read sequencing and provide insights on the evolutionary pattern and effect of SVs on morphological traits during rice domestication.
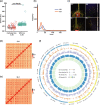
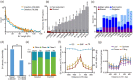
Results: In this study, we identified 318,589 SVs in cultivated and wild rice populations through a comprehensive analysis of 13 high-quality rice genomes and found that wild rice genomes contain 49% of unique SVs and an average of 1.76% of genes were lost during rice domestication. These SVs were further genotyped for 649 rice accessions, their evolutionary pattern during rice domestication and potential association with the diversity of important agronomic traits were examined. Genome-wide association studies between these SVs and nine agronomic traits identified 413 candidate causal variants, which together affect 361 genes. An 824-bp deletion in japonica rice, which encodes a serine carboxypeptidase family protein, is shown to be associated with grain length.
Conclusions: We provide relatively accurate and complete SV datasets for cultivated and wild rice accessions, especially in TE-rich regions, by comparing long-read sequencing data for 13 representative varieties. The integrated rice SV map and the identified candidate genes and variants represent valuable resources for future genomic research and breeding in rice.
Keywords: Association; Domestication; Oryza; Phenotype; Structure variation.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
-
- Li Y, Xiao J, Chen L, Huang X, Cheng Z, Han B, et al. Rice functional genomics research: past decade and future. Mol Plant. 2018;11(3):359–380. - PubMed
-
- Lye ZN, Purugganan MD. Copy number variation in domestication. Trends Plant Sci. 2019;24(4):352–365. - PubMed
-
- Qin P, Lu H, Du H, Wang H, Chen W, Chen Z, et al. Pan-genome analysis of 33 genetically diverse rice accessions reveals hidden genomic variations. Cell. 2021;184(13):3542–3558 e3516. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

