Integrative single-cell RNA-seq and ATAC-seq analysis of myogenic differentiation in pig
- PMID: 36726129
- PMCID: PMC9893630
- DOI: 10.1186/s12915-023-01519-z
Integrative single-cell RNA-seq and ATAC-seq analysis of myogenic differentiation in pig
Abstract
Background: Skeletal muscle development is a multistep process whose understanding is central in a broad range of fields and applications, from the potential medical value to human society, to its economic value associated with improvement of agricultural animals. Skeletal muscle initiates in the somites, with muscle precursor cells generated in the dermomyotome and dermomyotome-derived myotome before muscle differentiation ensues, a developmentally regulated process that is well characterized in model organisms. However, the regulation of skeletal muscle ontogeny during embryonic development remains poorly defined in farm animals, for instance in pig. Here, we profiled gene expression and chromatin accessibility in developing pig somites and myotomes at single-cell resolution.
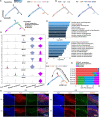
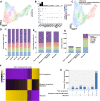
Results: We identified myogenic cells and other cell types and constructed a differentiation trajectory of pig skeletal muscle ontogeny. Along this trajectory, the dynamic changes in gene expression and chromatin accessibility coincided with the activities of distinct cell type-specific transcription factors. Some novel genes upregulated along the differentiation trajectory showed higher expression levels in muscular dystrophy mice than that in healthy mice, suggesting their involvement in myogenesis. Integrative analysis of chromatin accessibility, gene expression data, and in vitro experiments identified EGR1 and RHOB as critical regulators of pig embryonic myogenesis.
Conclusions: Collectively, our results enhance our understanding of the molecular and cellular dynamics in pig embryonic myogenesis and offer a high-quality resource for the further study of pig skeletal muscle development and human muscle disease.
Keywords: Myogenic differentiation; Pig; Skeletal muscle; scATAC-seq; scRNA-seq.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures







References
-
- Buckingham M, Rigby PWJ. Gene regulatory networks and transcriptional mechanisms that control myogenesis. Dev Cell. 2014;28:225–238. - PubMed
-
- Buckingham M, Montarras D, Relaix F, Cumano A, Mansouri A, Morgan J, et al. Pax3 and Pax7 mark a major population of muscle progenitor cells that contribute to skeletal muscle formation and regeneration. Neuromuscul Disord. 2006;16:S48–S48.
-
- Picard B, Berri C, Lefaucheur L, Molette C, Sayd T, Terlouw C. Skeletal muscle proteomics in livestock production. Brief Funct Genomics. 2010;9:259–278. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

