Anellovirus evolution during long-term chronic infection
- PMID: 36726484
- PMCID: PMC9885978
- DOI: 10.1093/ve/vead001
Anellovirus evolution during long-term chronic infection
Abstract
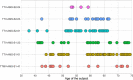
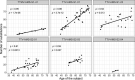
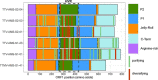
Human anelloviruses (AVs) are extremely genetically diverse, are widespread in the human population, and cause chronic infections. However, the evolutionary dynamics of AVs within single hosts is currently unknown, and it is unclear whether these changes have an implication on the long-term persistence of AVs in the host. Here, we assessed the evolutionary dynamics of six AV lineages during 30 years of chronic infection at single host resolution. The total number of substitutions and the number of variable sites increased over time. However, not all substitutions reached population fixation, showing that AV lineages form heterogeneous swarms within the host. Most substitutions occurred within a hypervariable region (HVR) located between nucleotide positions 800 and 1,300 of ORF1, which is known to be located within the spike domain. Different regions of the ORF1 gene undergo either positive or negative selection pressure. Sites under strong diversifying selection pressure were detected in the HVR, while the majority of the sites under purifying selection were detected outside this region. The HVR may play the role of an immunological decoy that prevents antibodies from binding to more vulnerable parts of ORF1. Moreover, the frequent substitutions in this region may increase the chances of AV particles escaping immune recognition.
Keywords: anellome; anellovirus; genetic variability; selection pressure; viral swarm; virus evolution.
© The Author(s) 2023. Published by Oxford University Press.
Figures


References
-
- Arze C. A. et al. (2021) ‘Global Genome Analysis Reveals a Vast and Dynamic Anellovirus Landscape within the Human Virome’, Cell Host & Microbe, 29: 1305–1315.e6. - PubMed
-
- Ball J. K. et al. (1999) ‘TT Virus Sequence Heterogeneity In Vivo: Evidence for Co-Infection with Multiple Genetic Types’, Journal of General Virology, 80: 1759–68. - PubMed
-
- Bédarida S. et al. (2017) ‘Analysis of Anelloviridae Sequences Characterized from Serial Human and Animal Biological Samples’, Infection, Genetics and Evolution, 53: 89–93. - PubMed
-
- Domingo E. et al. (1985) ‘The Quasispecies (Extremely Heterogeneous) Nature of Viral RNA Genome Populations: Biological Relevance—A Review’, Gene, 40: 1–8. - PubMed
LinkOut - more resources
Full Text Sources

