Statistical shape modeling of multi-organ anatomies with shared boundaries
- PMID: 36727040
- PMCID: PMC9886138
- DOI: 10.3389/fbioe.2022.1078800
Statistical shape modeling of multi-organ anatomies with shared boundaries
Abstract
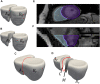
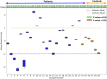
Introduction: Statistical shape modeling (SSM) is a valuable and powerful tool to generate a detailed representation of complex anatomy that enables quantitative analysis of shapes and their variations. SSM applies mathematics, statistics, and computing to parse the shape into some quantitative representation (such as correspondence points or landmarks) which can be used to study the covariance patterns of the shapes and answer various questions about the anatomical variations across the population. Complex anatomical structures have many diverse parts with varying interactions or intricate architecture. For example, the heart is a four-chambered organ with several shared boundaries between chambers. Subtle shape changes within the shared boundaries of the heart can indicate potential pathologic changes such as right ventricular overload. Early detection and robust quantification could provide insight into ideal treatment techniques and intervention timing. However, existing SSM methods do not explicitly handle shared boundaries which aid in a better understanding of the anatomy of interest. If shared boundaries are not explicitly modeled, it restricts the capability of the shape model to identify the pathological shape changes occurring at the shared boundary. Hence, this paper presents a general and flexible data-driven approach for building statistical shape models of multi-organ anatomies with shared boundaries that explicitly model contact surfaces. Methods: This work focuses on particle-based shape modeling (PSM), a state-of-art SSM approach for building shape models by optimizing the position of correspondence particles. The proposed PSM strategy for handling shared boundaries entails (a) detecting and extracting the shared boundary surface and contour (outline of the surface mesh/isoline) of the meshes of the two organs, (b) followed by a formulation for a correspondence-based optimization algorithm to build a multi-organ anatomy statistical shape model that captures morphological and alignment changes of individual organs and their shared boundary surfaces throughout the population. Results: We demonstrate the shared boundary pipeline using a toy dataset of parameterized shapes and a clinical dataset of the biventricular heart models. The shared boundary model for the cardiac biventricular data achieves consistent parameterization of the shared surface (interventricular septum) and identifies the curvature of the interventricular septum as pathological shape differences.
Keywords: biventricular cardiac MRI; interventricular septum; particle-based shape modeling; shared boundaries; statistical shape modeling.
Copyright © 2023 Iyer, Morris, Zenger, Karanth, Khan, Orkild, Korshak and Elhabian.
Conflict of interest statement
AM has equity interest in Marrek, Inc. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

