Identification of an unfolded protein response-related signature for predicting the prognosis of pancreatic ductal adenocarcinoma
- PMID: 36727081
- PMCID: PMC9885260
- DOI: 10.3389/fonc.2022.1060508
Identification of an unfolded protein response-related signature for predicting the prognosis of pancreatic ductal adenocarcinoma
Abstract
Background: Pancreatic ductal adenocarcinoma (PDAC) is a highly aggressive lethal malignancy. An effective prognosis prediction model is urgently needed for treatment optimization.
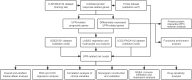
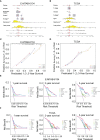
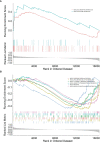
Methods: The differentially expressed unfolded protein response (UPR)‒related genes between pancreatic tumor and normal tissue were analyzed using the TCGA-PDAC dataset, and these genes that overlapped with UPR‒related prognostic genes from the E-MTAB-6134 dataset were further analyzed. Univariate, LASSO and multivariate Cox regression analyses were applied to establish a prognostic gene signature, which was evaluated by Kaplan‒Meier curve and receiver operating characteristic (ROC) analyses. E‒MTAB‒6134 was set as the training dataset, while TCGA-PDAC, GSE21501 and ICGC-PACA-AU were used for external validation. Subsequently, a nomogram integrating risk scores and clinical parameters was established, and gene set enrichment analysis (GSEA), tumor immunity analysis and drug sensitivity analysis were conducted.
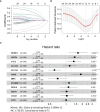
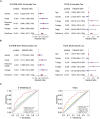
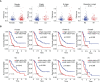
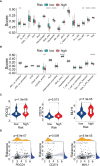
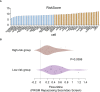
Results: A UPR-related signature comprising twelve genes was constructed and divided PDAC patients into high- and low-risk groups based on the median risk score. The UPR-related signature accurately predicted the prognosis and acted as an independent prognostic factor of PDAC patients, and the AUCs of the UPR-related signature in predicting PDAC prognosis at 1, 2 and 3 years were all more than 0.7 in the training and validation datasets. The UPR-related signature showed excellent performance in outcome prediction even in different clinicopathological subgroups, including the female (p<0.0001), male (p<0.0001), grade 1/2 (p<0.0001), grade 3 (p=0.028), N0 (p=0.043), N1 (p<0.001), and R0 (p<0.0001) groups. Furthermore, multiple immune-related pathways were enriched in the low-risk group, and risk scores in the low-risk group were also associated with significantly higher levels of tumor-infiltrating lymphocytes (TILs). In addition, DepMap drug sensitivity analysis and our validation experiment showed that PDAC cell lines with high UPR-related risk scores or UPR activation are more sensitive to floxuridine, which is used as an antineoplastic agent.
Conclusion: Herein, we identified a novel UPR-related prognostic signature that showed high value in predicting survival in patients with PDAC. Targeting these UPR-related genes might be an alternative for PDAC therapy. Further experimental studies are required to reveal how these genes mediate ER stress and PDAC progression.
Keywords: pancreatic cancer; prognostic model; risk score; survival analysis; unfolded protein response.
Copyright © 2023 Fang, Chen, Gong, Xia, Guan, Quan, Li, Zeng, Chen, Du and Liu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources

