MIMT1 and LINC01550 are uncharted lncRNAs down-regulated in colorectal cancer
- PMID: 36727289
- PMCID: PMC10182369
- DOI: 10.1111/iep.12467
MIMT1 and LINC01550 are uncharted lncRNAs down-regulated in colorectal cancer
Abstract
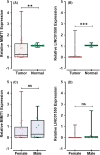
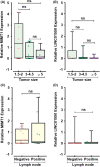
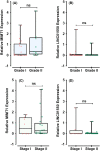
Incomplete knowledge of the molecular basis of colorectal cancer, with subsequent limitations in early diagnosis and effective treatment, has contributed to this form of malignancy becoming the second most common cause of cancer-related death worldwide. With the advances in high-throughput profiling techniques and the availability of public data sets such as The Cancer Genome Atlas Program (TCGA), a broad range of coding transcripts have been profiled and their underlying modes of action have been mapped. However, there is still a huge gap in our understanding of noncoding RNA dysregulation. To this end, we used a bioinformatics approach to shortlist and evaluate yet-to be-profiled long noncoding RNAs (lncRNAs) in colorectal cancer. We analysed the TCGA RNA-seq data and followed this by validating the expression patterns using a qPCR technique. Analysing in-house clinical samples, the real-time PCR method revealed that the shortlisted lncRNAs, that is MER1 Repeat Containing Imprinted Transcript 1 (MIMT1) and Non-Protein Coding RNA 1550 (LINC01550), were down-regulated in colorectal cancer tumours compared with the paired adjacent normal tissues. Mechanistically, the in silico results suggest that LINC01550 could form a complex competitive endogenous RNA (ceRNA) network leading to the subsequent regulation of colorectal cancer-related genes, such as CUGBP Elav-Like Family Member (CELF2), Polypyrimidine Tract Binding Protein 1 (PTBP1) and ELAV Like RNA Binding Protein 1 (ELAV1). The findings of this work indicate that MIMT1 and LINC01550 could be novel tumour suppressor genes that can be studied further to assess their roles in regulating the cancer signalling pathway(s).
Keywords: LINC01550; MIMT1; ceRNA; colorectal cancer; long noncoding RNA.
© 2023 Company of the International Journal of Experimental Pathology (CIJEP).
Conflict of interest statement
All authors declare that they have no conflict of interest.
Figures
References
-
- Xiao W, Qu X, Li X, et al. Identification of commonly dysregulated genes in colorectal cancer by integrating analysis of RNA‐seq data and qRT‐PCR validation. Cancer Gene Ther. 2015;22(5):278‐284. - PubMed
-
- Nannini M, Pantaleo MA, Maleddu A, Astolfi A, Formica S, Biasco G. Gene expression profiling in colorectal cancer using microarray technologies: results and perspectives. Cancer Treat Rev. 2009;35(3):201‐209. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

