SMAD3 promotes expression and activity of the androgen receptor in prostate cancer
- PMID: 36727462
- PMCID: PMC10085708
- DOI: 10.1093/nar/gkad043
SMAD3 promotes expression and activity of the androgen receptor in prostate cancer
Abstract
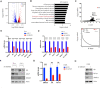
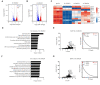
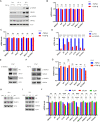
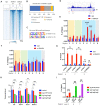
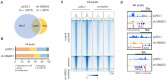
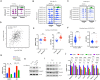
Overexpression of androgen receptor (AR) is the primary cause of castration-resistant prostate cancer, although mechanisms upregulating AR transcription in this context are not well understood. Our RNA-seq studies revealed that SMAD3 knockdown decreased levels of AR and AR target genes, whereas SMAD4 or SMAD2 knockdown had little or no effect. ChIP-seq analysis showed that SMAD3 knockdown decreased global binding of AR to chromatin. Mechanistically, we show that SMAD3 binds to intron 3 of the AR gene to promote AR expression. Targeting these binding sites by CRISPRi reduced transcript levels of AR and AR targets. In addition, ∼50% of AR and SMAD3 ChIP-seq peaks overlapped, and SMAD3 may also cooperate with or co-activate AR for AR target expression. Functionally, AR re-expression in SMAD3-knockdown cells partially rescued AR target expression and cell growth defects. The SMAD3 peak in AR intron 3 overlapped with H3K27ac ChIP-seq and ATAC-seq peaks in datasets of prostate cancer. AR and SMAD3 mRNAs were upregulated in datasets of metastatic prostate cancer and CRPC compared with primary prostate cancer. A SMAD3 PROTAC inhibitor reduced levels of AR, AR-V7 and AR targets in prostate cancer cells. This study suggests that SMAD3 could be targeted to inhibit AR in prostate cancer.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Fizazi K., Scher H.I., Molina A., Logothetis C.J., Chi K.N., Jones R.J., Staffurth J.N., North S., Vogelzang N.J., Saad F.et al.. Abiraterone acetate for treatment of metastatic castration-resistant prostate cancer: final overall survival analysis of the COU-AA-301 randomised, double-blind, placebo-controlled phase 3 study. Lancet Oncol. 2012; 13:983–992. - PubMed
-
- Chen C.D., Welsbie D.S., Tran C., Baek S.H., Chen R., Vessella R., Rosenfeld M.G., Sawyers C.L.. Molecular determinants of resistance to antiandrogen therapy. Nat. Med. 2004; 10:33–39. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

