Arginase 1 is a key driver of immune suppression in pancreatic cancer
- PMID: 36727849
- PMCID: PMC10260021
- DOI: 10.7554/eLife.80721
Arginase 1 is a key driver of immune suppression in pancreatic cancer
Abstract
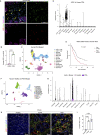
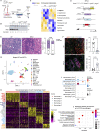
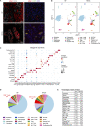
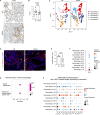
An extensive fibroinflammatory stroma rich in macrophages is a hallmark of pancreatic cancer. In this disease, it is well appreciated that macrophages are immunosuppressive and contribute to the poor response to immunotherapy; however, the mechanisms of immune suppression are complex and not fully understood. Immunosuppressive macrophages are classically defined by the expression of the enzyme Arginase 1 (ARG1), which we demonstrated is potently expressed in pancreatic tumor-associated macrophages from both human patients and mouse models. While routinely used as a polarization marker, ARG1 also catabolizes arginine, an amino acid required for T cell activation and proliferation. To investigate this metabolic function, we used a genetic and a pharmacologic approach to target Arg1 in pancreatic cancer. Genetic inactivation of Arg1 in macrophages, using a dual recombinase genetically engineered mouse model of pancreatic cancer, delayed formation of invasive disease, while increasing CD8+ T cell infiltration. Additionally, Arg1 deletion induced compensatory mechanisms, including Arg1 overexpression in epithelial cells, namely Tuft cells, and Arg2 overexpression in a subset of macrophages. To overcome these compensatory mechanisms, we used a pharmacological approach to inhibit arginase. Treatment of established tumors with the arginase inhibitor CB-1158 exhibited further increased CD8+ T cell infiltration, beyond that seen with the macrophage-specific knockout, and sensitized the tumors to anti-PD1 immune checkpoint blockade. Our data demonstrate that Arg1 drives immune suppression in pancreatic cancer by depleting arginine and inhibiting T cell activation.
Keywords: CD8 T cells; PDA; arginase; cancer biology; human; immunosuppression; immunotherapy; mouse; myeloid.
© 2023, Menjivar et al.
Conflict of interest statement
RM, ZN, WD, KD, HH, CE, KB, AV, WY, FL, AB, PK, DS, HC, FB, EC, YZ, CH, MP No competing interests declared, CL has received consulting fees from Astellas Pharmaceuticals, Odyssey Therapeutics, and T-Knife Therapeutics, and is an inventor on patents pertaining to Kras regulated metabolic pathways, redox control pathways in pancreatic cancer, and targeting the GOT1-pathway as a therapeutic approach (US Patent No: 2015126580-A1, 05/07/2015; US Patent No: 20190136238, 05/09/2019; International Patent No: WO2013177426-A2, 04/23/2015)
Figures





Comment in
-
How nearby nutrients shape tumor growth.Elife. 2023 Jul 17;12:e89825. doi: 10.7554/eLife.89825. Elife. 2023. PMID: 37459171 Free PMC article.
References
-
- Balachandran VP, Łuksza M, Zhao JN, Makarov V, Moral JA, Remark R, Herbst B, Askan G, Bhanot U, Senbabaoglu Y, Wells DK, Cary CIO, Grbovic-Huezo O, Attiyeh M, Medina B, Zhang J, Loo J, Saglimbeni J, Abu-Akeel M, Zappasodi R, Riaz N, Smoragiewicz M, Kelley ZL, Basturk O, Gönen M, Levine AJ, Allen PJ, Fearon DT, Merad M, Gnjatic S, Iacobuzio-Donahue CA, Wolchok JD, DeMatteo RP, Chan TA, Greenbaum BD, Merghoub T, Leach SD, Australian Pancreatic Cancer Genome Initiative. Garvan Institute of Medical Research. Prince of Wales Hospital. Royal North Shore Hospital. University of Glasgow. St Vincent’s Hospital. QIMR Berghofer Medical Research Institute. University of Melbourne, Centre for Cancer Research. University of Queensland, Institute for Molecular Bioscience. Bankstown Hospital. Liverpool Hospital. Royal Prince Alfred Hospital, Chris O’Brien Lifehouse. Westmead Hospital. Fremantle Hospital. St John of God Healthcare. Royal Adelaide Hospital. Flinders Medical Centre. Envoi Pathology. Princess Alexandria Hospital. Austin Hospital. Johns Hopkins Medical Institutes. ARC-Net Centre for Applied Research on Cancer Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature. 2017;551:512–516. doi: 10.1038/nature24462. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
- K99 CA267176/CA/NCI NIH HHS/United States
- P30 CA046592/CA/NCI NIH HHS/United States
- F32 CA228328/CA/NCI NIH HHS/United States
- T32 GM145304/GM/NIGMS NIH HHS/United States
- R01 CA248160/CA/NCI NIH HHS/United States
- S10 OD028612/OD/NIH HHS/United States
- U01 CA274154/CA/NCI NIH HHS/United States
- U01 CA224145/CA/NCI NIH HHS/United States
- T32 GM007315/GM/NIGMS NIH HHS/United States
- F31 CA257533/CA/NCI NIH HHS/United States
- T32 GM008353/GM/NIGMS NIH HHS/United States
- R01 CA198074/CA/NCI NIH HHS/United States
- R00 CA241357/CA/NCI NIH HHS/United States
- T32 HD007505/HD/NICHD NIH HHS/United States
- R01 CA151588/CA/NCI NIH HHS/United States
- R25 GM086262/GM/NIGMS NIH HHS/United States
- T32 CA009676/CA/NCI NIH HHS/United States
- R01 CA244931/CA/NCI NIH HHS/United States
- F31 CA247037/CA/NCI NIH HHS/United States
- P30 CA062203/CA/NCI NIH HHS/United States
- R01 CA247516/CA/NCI NIH HHS/United States
- T32 DK094775/DK/NIDDK NIH HHS/United States
- T32 AI007413/AI/NIAID NIH HHS/United States
- R50 CA232985/CA/NCI NIH HHS/United States
- R37 CA237421/CA/NCI NIH HHS/United States
- R01 CA268426/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

