Siglec-9 Restrains Antibody-Dependent Natural Killer Cell Cytotoxicity against SARS-CoV-2
- PMID: 36728420
- PMCID: PMC9973332
- DOI: 10.1128/mbio.03393-22
Siglec-9 Restrains Antibody-Dependent Natural Killer Cell Cytotoxicity against SARS-CoV-2
Abstract
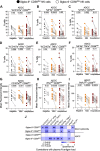
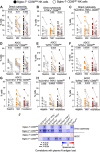
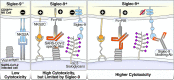
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection alters the immunological profiles of natural killer (NK) cells. However, whether NK antiviral functions are impaired during severe coronavirus disease 2019 (COVID-19) and what host factors modulate these functions remain unclear. We found that NK cells from hospitalized COVID-19 patients degranulate less against SARS-CoV-2 antigen-expressing cells (in direct cytolytic and antibody-dependent cell cytotoxicity [ADCC] assays) than NK cells from mild COVID-19 patients or negative controls. The lower NK degranulation was associated with higher plasma levels of SARS-CoV-2 nucleocapsid antigen. Phenotypic and functional analyses showed that NK cells expressing the glyco-immune checkpoint Siglec-9 elicited higher ADCC than Siglec-9- NK cells. Consistently, Siglec-9+ NK cells exhibit an activated and mature phenotype with higher expression of CD16 (FcγRIII; mediator of ADCC), CD57 (maturation marker), and NKG2C (activating receptor), along with lower expression of the inhibitory receptor NKG2A, than Siglec-9- CD56dim NK cells. These data are consistent with the concept that the NK cell subpopulation expressing Siglec-9 is highly activated and cytotoxic. However, the Siglec-9 molecule itself is an inhibitory receptor that restrains NK cytotoxicity during cancer and other viral infections. Indeed, blocking Siglec-9 significantly enhanced the ADCC-mediated NK degranulation and lysis of SARS-CoV-2-antigen-positive target cells. These data support a model in which the Siglec-9+ CD56dim NK subpopulation is cytotoxic even while it is restrained by the inhibitory effects of Siglec-9. Alleviating the Siglec-9-mediated restriction on NK cytotoxicity may further improve NK immune surveillance and presents an opportunity to develop novel immunotherapeutic tools against SARS-CoV-2 infected cells. IMPORTANCE One mechanism that cancer cells use to evade natural killer cell immune surveillance is by expressing high levels of sialoglycans, which bind to Siglec-9, a glyco-immune checkpoint molecule on NK cells. This binding inhibits NK cell cytotoxicity. Several viruses, such as hepatitis B virus (HBV) and HIV, also use a similar mechanism to evade NK surveillance. We found that NK cells from SARS-CoV-2-hospitalized patients are less able to function against cells expressing SARS-CoV-2 Spike protein than NK cells from SARS-CoV-2 mild patients or uninfected controls. We also found that the cytotoxicity of the Siglec-9+ NK subpopulation is indeed restrained by the inhibitory nature of the Siglec-9 molecule and that blocking Siglec-9 can enhance the ability of NK cells to target cells expressing SARS-CoV-2 antigens. Our results suggest that a targetable glyco-immune checkpoint mechanism, Siglec-9/sialoglycan interaction, may contribute to the ability of SARS-CoV-2 to evade NK immune surveillance.
Keywords: COVID-19; SARS-CoV-2; Siglec-7; Siglec-9; antibody-dependent cell cytotoxicity; natural killer cells.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, Liu L, Shan H, Lei CL, Hui DSC, Du B, Li LJ, Zeng G, Yuen KY, Chen RC, Tang CL, Wang T, Chen PY, Xiang J, Li SY, Wang JL, Liang ZJ, Peng YX, Wei L, Liu Y, Hu YH, Peng P, Wang JM, Liu JY, Chen Z, Li G, Zheng ZJ, Qiu SQ, Luo J, Ye CJ, Zhu SY, Zhong NS, China Medical Treatment Expert Group for Covid-19 . 2020. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med 382:1708–1720. doi:10.1056/NEJMoa2002032. - DOI - PMC - PubMed
-
- Schulien I, Kemming J, Oberhardt V, Wild K, Seidel LM, Killmer S, Sagar Daul F, Salvat Lago M, Decker A, Luxenburger H, Binder B, Bettinger D, Sogukpinar O, Rieg S, Panning M, Huzly D, Schwemmle M, Kochs G, Waller CF, Nieters A, Duerschmied D, Emmerich F, Mei HE, Schulz AR, Llewellyn-Lacey S, Price DA, Boettler T, Bengsch B, Thimme R, Hofmann M, Neumann-Haefelin C. 2021. Characterization of pre-existing and induced SARS-CoV-2-specific CD8+ T cells. Nat Med 27:78–85. doi:10.1038/s41591-020-01143-2. - DOI - PubMed
-
- Stephenson E, Reynolds G, Botting RA, Calero-Nieto FJ, Morgan MD, Tuong ZK, Bach K, Sungnak W, Worlock KB, Yoshida M, Kumasaka N, Kania K, Engelbert J, Olabi B, Spegarova JS, Wilson NK, Mende N, Jardine L, Gardner LCS, Goh I, Horsfall D, McGrath J, Webb S, Mather MW, Lindeboom RGH, Dann E, Huang N, Polanski K, Prigmore E, Gothe F, Scott J, Payne RP, Baker KF, Hanrath AT, Schim van der Loeff ICD, Barr AS, Sanchez-Gonzalez A, Bergamaschi L, Mescia F, Barnes JL, Kilich E, de Wilton A, Saigal A, Saleh A, Janes SM, Smith CM, Gopee N, Wilson C, Coupland P, Coxhead JM, Cambridge Institute of Therapeutic Immunology and Infectious Disease-National Institute of Health Research (CITIID-NIHR) COVID-19 BioResource Collaboration , et al. 2021. Single-cell multi-omics analysis of the immune response in COVID-19. Nat Med 27:904–916. doi:10.1038/s41591-021-01329-2. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

