Effect of ATG12-ATG5-ATG16L1 autophagy E3-like complex on the ability of LC3/GABARAP proteins to induce vesicle tethering and fusion
- PMID: 36729310
- PMCID: PMC9894987
- DOI: 10.1007/s00018-023-04704-z
Effect of ATG12-ATG5-ATG16L1 autophagy E3-like complex on the ability of LC3/GABARAP proteins to induce vesicle tethering and fusion
Abstract
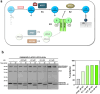
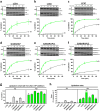
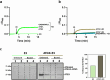
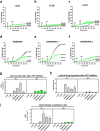
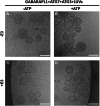
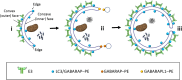
In macroautophagy, the autophagosome (AP) engulfs portions of cytoplasm to allow their lysosomal degradation. AP formation in humans requires the concerted action of the ATG12 and LC3/GABARAP conjugation systems. The ATG12-ATG5-ATG16L1 or E3-like complex (E3 for short) acts as a ubiquitin-like E3 enzyme, promoting LC3/GABARAP proteins anchoring to the AP membrane. Their role in the AP expansion process is still unclear, in part because there are no studies comparing six LC3/GABARAP family member roles under the same conditions, and also because the full human E3 was only recently available. In the present study, the lipidation of six members of the LC3/GABARAP family has been reconstituted in the presence and absence of E3, and the mechanisms by which E3 and LC3/GABARAP proteins participate in vesicle tethering and fusion have been investigated. In the absence of E3, GABARAP and GABARAPL1 showed the highest activities. Differences found within LC3/GABARAP proteins suggest the existence of a lipidation threshold, lower for the GABARAP subfamily, as a requisite for tethering and inter-vesicular lipid mixing. E3 increases and speeds up lipidation and LC3/GABARAP-promoted tethering. However, E3 hampers LC3/GABARAP capacity to induce inter-vesicular lipid mixing or subsequent fusion, presumably through the formation of a rigid scaffold on the vesicle surface. Our results suggest a model of AP expansion in which the growing regions would be areas where the LC3/GABARAP proteins involved should be susceptible to lipidation in the absence of E3, or else a regulatory mechanism would allow vesicle incorporation and phagophore growth when E3 is present.
Keywords: ATG12 UBL system; Autophagosome expansion; Autophagy conjugation systems; Human ATG8; Lipid-protein interaction; Membrane fusion.
© 2023. The Author(s).
Conflict of interest statement
The authors have not disclosed any competing interests.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

