De Novo Mutations Contributes Approximately 7% of Pathogenicity in Inherited Eye Diseases
- PMID: 36729443
- PMCID: PMC9907368
- DOI: 10.1167/iovs.64.2.5
De Novo Mutations Contributes Approximately 7% of Pathogenicity in Inherited Eye Diseases
Abstract
Purpose: The purpose of this study was to describe genotype-phenotype associations and novel insights into genetic characteristics in a trio-based cohort of inherited eye diseases (IEDs).
Methods: To determine the etiological role of de novo mutations (DNMs) and genetic profile in IEDs, we retrospectively reviewed a large cohort of proband-parent trios of Chinese origin. The patients underwent a detailed examination and was clinically diagnosed by an ophthalmologist. Panel-based targeted exome sequencing was performed on DNA extracted from blood samples, containing coding regions of 792 IED-causative genes and their flanking exons. All participants underwent genetic testing.
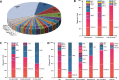
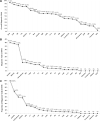
Results: All proband-parent trios were divided into 22 subgroups, the overall diagnostic yield was 48.67% (605/1243), ranging from 4% to 94.44% for each of the subgroups. A total of 108 IED-causative genes were identified, with the top 24 genes explaining 67% of the 605 genetically solved trios. The genetic etiology of 6.76% (84/1243) of the trio was attributed to disease-causative DNMs, and the top 3 subgroups with the highest incidence of DNM were aniridia (n = 40%), Marfan syndrome/ectopia lentis (n = 38.78%), and retinoblastoma (n = 37.04%). The top 10 genes have a diagnostic yield of DNM greater than 3.5% in their subgroups, including PAX6 (40.00%), FBN1 (38.78%), RB1 (37.04%), CRX (10.34%), CHM (9.09%), WFS1 (8.00%), RP1L1 (5.88%), RS1 (5.26%), PCDH15 (4.00%), and ABCA4 (3.51%). Additionally, the incidence of DNM in offspring showed a trend of correlation with paternal age at reproduction, but not statistically significant with paternal (P = 0.154) and maternal (P = 0.959) age at reproduction.
Conclusions: Trios-based genetic analysis has high accuracy and validity. Our study helps to quantify the burden of the full spectrum IED caused by each gene, offers novel potential for elucidating etiology, and plays a crucial role in genetic counseling and patient management.
Conflict of interest statement
Disclosure:
Figures


References
-
- Solebo AL, Teoh L, Rahi J.. Epidemiology of blindness in children. Arch Dis Child. 2017; 102: 853–857. - PubMed
-
- Wygnanski-Jaffe T, Levin AV.. Introductory genetics for the ophthalmologist. Am Acad Ophthalmol, Focal Points. Clin Module Ophthalmol. 2005; 23: 1–11.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

