Integrative miRNA-mRNA profiling of human epidermis: unique signature of SCN9A painful neuropathy
- PMID: 36730021
- PMCID: PMC10316770
- DOI: 10.1093/brain/awad025
Integrative miRNA-mRNA profiling of human epidermis: unique signature of SCN9A painful neuropathy
Abstract
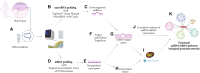
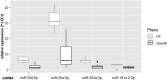
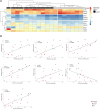
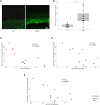
Personalized management of neuropathic pain is an unmet clinical need due to heterogeneity of the underlying aetiologies, incompletely understood pathophysiological mechanisms and limited efficacy of existing treatments. Recent studies on microRNA in pain preclinical models have begun to yield insights into pain-related mechanisms, identifying nociception-related species differences and pinpointing potential drug candidates. With the aim of bridging the translational gap towards the clinic, we generated a human pain-related integrative miRNA and mRNA molecular profile of the epidermis, the tissue hosting small nerve fibres, in a deeply phenotyped cohort of patients with sodium channel-related painful neuropathy not responding to currently available therapies. We identified four miRNAs strongly discriminating patients from healthy individuals, confirming their effect on differentially expressed gene targets driving peripheral sensory transduction, transmission, modulation and post-transcriptional modifications, with strong effects on gene targets including NEDD4. We identified a complex epidermal miRNA-mRNA network based on tissue-specific experimental data suggesting a cross-talk between epidermal cells and axons in neuropathy pain. Using immunofluorescence assay and confocal microscopy, we observed that Nav1.7 signal intensity in keratinocytes strongly inversely correlated with NEDD4 expression that was downregulated by miR-30 family, suggesting post-transcriptional fine tuning of pain-related protein expression. Our targeted molecular profiling advances the understanding of specific neuropathic pain fine signatures and may accelerate process towards personalized medicine in patients with neuropathic pain.
Keywords: SCN9A; miRNA; neuropathic pain; skin biopsy; small fibre neuropathy.
© The Author(s) 2023. Published by Oxford University Press on behalf of the Guarantors of Brain.
Conflict of interest statement
The authors report no competing interests.
Figures

References
-
- Baron R, Binder A, Wasner G. Neuropathic pain: Diagnosis, pathophysiological mechanisms, and treatment. Lancet Neurol. 2010;9:807–819. - PubMed
-
- Bouhassira D. Neuropathic pain: Definition, assessment and epidemiology. Rev Neurol (Paris). 2019;175(1–2):16–25. - PubMed
-
- van Hecke O, Austin SK, Khan RA, Smith BH, Torrance N. Neuropathic pain in the general population: A systematic review of epidemiological studies. Pain. 2014;155:654–662. - PubMed

