Spike Gene Target Amplification in a Diagnostic Assay as a Marker for Public Health Monitoring of Emerging SARS-CoV-2 Variants - United States, November 2021-January 2023
- PMID: 36730050
- PMCID: PMC9927069
- DOI: 10.15585/mmwr.mm7205e2
Spike Gene Target Amplification in a Diagnostic Assay as a Marker for Public Health Monitoring of Emerging SARS-CoV-2 Variants - United States, November 2021-January 2023
Abstract
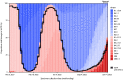
Monitoring emerging SARS-CoV-2 lineages and their epidemiologic characteristics helps to inform public health decisions regarding vaccine policy, the use of therapeutics, and health care capacity. When the SARS-CoV-2 Alpha variant emerged in late 2020, a spike gene (S-gene) deletion (Δ69-70) in the N-terminal region, which might compensate for immune escape mutations that impair infectivity (1), resulted in reduced or failed S-gene target amplification in certain multitarget reverse transcription-polymerase chain reaction (RT-PCR) assays, a pattern referred to as S-gene target failure (SGTF) (2). The predominant U.S. SARS-CoV-2 lineages have generally alternated between SGTF and S-gene target presence (SGTP), which alongside genomic sequencing, has facilitated early monitoring of emerging variants. During a period when Omicron BA.5-related sublineages (which exhibit SGTF) predominated, an XBB.1.5 sublineage with SGTP has rapidly expanded in the northeastern United States and other regions.
Conflict of interest statement
All authors have completed and submitted the International Committee of Medical Journal Editors form for disclosure of potential conflicts of interest. No other potential conflicts of interest were disclosed.
Figures
References
-
- Lambrou AS, Shirk P, Steele MK, et al. ; Strain Surveillance and Emerging Variants Bioinformatic Working Group; Strain Surveillance and Emerging Variants NS3 Working Group. Genomic surveillance for SARS-CoV-2 variants: predominance of the Delta (B.1.617.2) and Omicron (B.1.1.529) variants—United States, June 2021–January 2022. MMWR Morb Mortal Wkly Rep 2022;71:206–11. 10.15585/mmwr.mm7106a4 - DOI - PMC - PubMed
-
- Link-Gelles R, Ciesla AA, Roper LE, et al. Early estimates of bivalent mRNA booster dose vaccine effectiveness in preventing symptomatic SARS-CoV-2 infection attributable to Omicron BA.5- and XBB/XBB.1.5-related sublineages among immunocompetent adults—Increasing Community Access to Testing Program, United States, December 2022–January 2023. MMWR Morb Mortal Wkly Rep 2023;72. https://www.cdc.gov/mmwr/volumes/72/wr/mm7205e2.htm?s_cid=mm7205e2_w - PMC - PubMed
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

