Breakdown of clonal cooperative architecture in multispecies biofilms and the spatial ecology of predation
- PMID: 36730197
- PMCID: PMC9963355
- DOI: 10.1073/pnas.2212650120
Breakdown of clonal cooperative architecture in multispecies biofilms and the spatial ecology of predation
Abstract
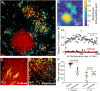
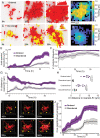
Biofilm formation, including adherence to surfaces and secretion of extracellular matrix, is common in the microbial world, but we often do not know how interaction at the cellular spatial scale translates to higher-order biofilm community ecology. Here we explore an especially understudied element of biofilm ecology, namely predation by the bacterium Bdellovibrio bacteriovorus. This predator can kill and consume many different Gram-negative bacteria, including Vibrio cholerae and Escherichia coli. V. cholerae can protect itself from predation within densely packed biofilm structures that it creates, whereas E. coli biofilms are highly susceptible to B. bacteriovorus. We explore how predator-prey dynamics change when V. cholerae and E. coli are growing in biofilms together. We find that in dual-species prey biofilms, E. coli survival under B. bacteriovorus predation increases, whereas V. cholerae survival decreases. E. coli benefits from predator protection when it becomes embedded within expanding groups of highly packed V. cholerae. But we also find that the ordered, highly packed, and clonal biofilm structure of V. cholerae can be disrupted if V. cholerae cells are directly adjacent to E. coli cells at the start of biofilm growth. When this occurs, the two species become intermixed, and the resulting disordered cell groups do not block predator entry. Because biofilm cell group structure depends on initial cell distributions at the start of prey biofilm growth, the surface colonization dynamics have a dramatic impact on the eventual multispecies biofilm architecture, which in turn determines to what extent both species survive exposure to B. bacteriovorus.
Keywords: architecture; biofilm; cooperation; matrix; predator–prey.
Conflict of interest statement
The authors declare no competing interest.
Figures



Comment in
-
Community outcomes depend on cooperative biofilm structure.Proc Natl Acad Sci U S A. 2023 Feb 7;120(6):e2221624120. doi: 10.1073/pnas.2221624120. Epub 2023 Feb 2. Proc Natl Acad Sci U S A. 2023. PMID: 36730195 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

