A cellular taxonomy of the adult human spinal cord
- PMID: 36731429
- PMCID: PMC10044516
- DOI: 10.1016/j.neuron.2023.01.007
A cellular taxonomy of the adult human spinal cord
Abstract
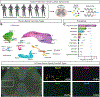
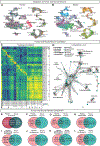
The mammalian spinal cord functions as a community of cell types for sensory processing, autonomic control, and movement. While animal models have advanced our understanding of spinal cellular diversity, characterizing human biology directly is important to uncover specialized features of basic function and human pathology. Here, we present a cellular taxonomy of the adult human spinal cord using single-nucleus RNA sequencing with spatial transcriptomics and antibody validation. We identified 29 glial clusters and 35 neuronal clusters, organized principally by anatomical location. To demonstrate the relevance of this resource to human disease, we analyzed spinal motoneurons, which degenerate in amyotrophic lateral sclerosis (ALS) and other diseases. We found that compared with other spinal neurons, human motoneurons are defined by genes related to cell size, cytoskeletal structure, and ALS, suggesting a specialized molecular repertoire underlying their selective vulnerability. We include a web resource to facilitate further investigations into human spinal cord biology.
Keywords: ALS; cell types; human spinal cord; motoneuron; single cell sequencing; spatial transcriptomics.
Published by Elsevier Inc.
Conflict of interest statement
Declaration of interests M.A.N.’s participation in this project was part of a competitive contract awarded to Data Tecnica International LLC by the National Institutes of Health to support open science research. He also currently serves on the scientific advisory board for Clover Therapeutics and is an advisor to Neuron23 Inc.
Figures





References
-
- Ravits J, Appel S, Baloh RH, Barohn R, Brooks BR, Elman L, Floeter MK, Henderson C, Lomen-Hoerth C, Macklis JD, et al. (2013). Deciphering amyotrophic lateral sclerosis: what phenotype, neuropathology and genetics are telling us about pathogenesis. Amyotroph Lateral Scler Frontotemporal Degener 14 Suppl 1, 5–18. 10.3109/21678421.2013.778548. - DOI - PMC - PubMed
-
- Blum JA, Klemm S, Shadrach JL, Guttenplan KA, Nakayama L, Kathiria A, Hoang PT, Gautier O, Kaltschmidt JA, Greenleaf WJ, and Gitler AD (2021). Single-cell transcriptomic analysis of the adult mouse spinal cord reveals molecular diversity of autonomic and skeletal motor neurons. Nat Neurosci 24, 572–583. 10.1038/s41593-020-00795-0. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

