Plant mobile domain proteins ensure Microrchidia 1 expression to fulfill transposon silencing
- PMID: 36732020
- PMCID: PMC9899485
- DOI: 10.26508/lsa.202201539
Plant mobile domain proteins ensure Microrchidia 1 expression to fulfill transposon silencing
Abstract
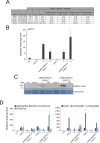
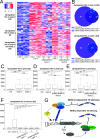
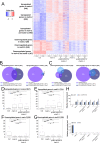
Silencing of transposable elements (TEs) is an essential process to maintain genomic integrity within the cell. In Arabidopsis, together with canonical epigenetic pathways such as DNA methylation and modifications of histone tails, the plant mobile domain (PMD) proteins MAINTENANCE OF MERISTEMS (MAIN) and MAIN-LIKE 1 (MAIL1) are involved in TE silencing. In addition, the MICRORCHIDIA (MORC) ATPases, including MORC1, are important cellular factors repressing TEs. Here, we describe the genetic interaction and connection between the PMD and MORC pathways by showing that MORC1 expression is impaired in main and mail1 mutants. Transcriptomic analyses of higher order mutant plants combining pmd and morc1 mutations, and pmd mutants in which MORC1 expression is restored, show that the silencing defects of a subset of TEs in pmd mutants are most likely the consequence of MORC1 down-regulation. Besides, a significant fraction of up-regulated TEs in pmd mutants are not targeted by the MORC1 pathway.
© 2023 Jarry et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

Similar articles
-
The plant mobile domain proteins MAIN and MAIL1 interact with the phosphatase PP7L to regulate gene expression and silence transposable elements in Arabidopsis thaliana.PLoS Genet. 2020 Apr 14;16(4):e1008324. doi: 10.1371/journal.pgen.1008324. eCollection 2020 Apr. PLoS Genet. 2020. PMID: 32287271 Free PMC article.
-
MORC family ATPases required for heterochromatin condensation and gene silencing.Science. 2012 Jun 15;336(6087):1448-51. doi: 10.1126/science.1221472. Epub 2012 May 3. Science. 2012. PMID: 22555433 Free PMC article.
-
Arabidopsis proteins with a transposon-related domain act in gene silencing.Nat Commun. 2017 May 3;8:15122. doi: 10.1038/ncomms15122. Nat Commun. 2017. PMID: 28466841 Free PMC article.
-
Control of transposable elements in Arabidopsis thaliana.Chromosome Res. 2014 Jun;22(2):217-23. doi: 10.1007/s10577-014-9417-9. Chromosome Res. 2014. PMID: 24801341 Review.
-
Silencing of transposons in plant genomes: kick them when they're down.Genome Biol. 2004;5(12):249. doi: 10.1186/gb-2004-5-12-249. Epub 2004 Nov 16. Genome Biol. 2004. PMID: 15575975 Free PMC article. Review.
Cited by
-
OsMAINTENANCE OF MERISTEM LIKE 1 controls style number at high temperatures in rice.Plant Mol Biol. 2025 Jan 21;115(1):24. doi: 10.1007/s11103-025-01553-1. Plant Mol Biol. 2025. PMID: 39836296
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
