WDR5 represents a therapeutically exploitable target for cancer stem cells in glioblastoma
- PMID: 36732025
- PMCID: PMC10069451
- DOI: 10.1101/gad.349803.122
WDR5 represents a therapeutically exploitable target for cancer stem cells in glioblastoma
Abstract
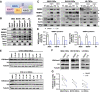
Glioblastomas (GBMs) are heterogeneous, treatment-resistant tumors driven by populations of cancer stem cells (CSCs). However, few molecular mechanisms critical for CSC population maintenance have been exploited for therapeutic development. We developed a spatially resolved loss-of-function screen in GBM patient-derived organoids to identify essential epigenetic regulators in the SOX2-enriched, therapy-resistant niche and identified WDR5 as indispensable for this population. WDR5 is a component of the WRAD complex, which promotes SET1 family-mediated Lys4 methylation of histone H3 (H3K4me), associated with positive regulation of transcription. In GBM CSCs, WDR5 inhibitors blocked WRAD complex assembly and reduced H3K4 trimethylation and expression of genes involved in CSC-relevant oncogenic pathways. H3K4me3 peaks lost with WDR5 inhibitor treatment occurred disproportionally on POU transcription factor motifs, including the POU5F1(OCT4)::SOX2 motif. Use of a SOX2/OCT4 reporter demonstrated that WDR5 inhibitor treatment diminished cells with high reporter activity. Furthermore, WDR5 inhibitor treatment and WDR5 knockdown altered the stem cell state, disrupting CSC in vitro growth and self-renewal, as well as in vivo tumor growth. These findings highlight the role of WDR5 and the WRAD complex in maintaining the CSC state and provide a rationale for therapeutic development of WDR5 inhibitors for GBM and other advanced cancers.
Keywords: epigenetics; glioblastoma; stem cell.
© 2023 Mitchell et al.; Published by Cold Spring Harbor Laboratory Press.
Figures






References
-
- Alicea-Velázquez NL, Shinsky SA, Loh DM, Lee JH, Skalnik DG, Cosgrove MS. 2016. Targeted disruption of the interaction between WD-40 repeat protein 5 (WDR5) and mixed lineage leukemia (MLL)/SET1 family proteins specifically inhibits MLL1 and SETd1A methyltransferase complexes. J Biol Chem 291: 22357–22372. 10.1074/jbc.M116.752626 - DOI - PMC - PubMed
-
- Alvarado AG, Thiagarajan PS, Mulkearns-Hubert EE, Silver DJ, Hale JS, Alban TJ, Turaga SM, Jarrar A, Reizes O, Longworth MS, et al. 2017. Glioblastoma cancer stem cells evade innate immune suppression of self-renewal through reduced TLR4 expression. Cell Stem Cell 20: 450–461.e4. 10.1016/j.stem.2016.12.001 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
