Phosphoinositide acyl chain saturation drives CD8+ effector T cell signaling and function
- PMID: 36732424
- PMCID: PMC10908374
- DOI: 10.1038/s41590-023-01419-y
Phosphoinositide acyl chain saturation drives CD8+ effector T cell signaling and function
Abstract
How lipidome changes support CD8+ effector T (Teff) cell differentiation is not well understood. Here we show that, although naive T cells are rich in polyunsaturated phosphoinositides (PIPn with 3-4 double bonds), Teff cells have unique PIPn marked by saturated fatty acyl chains (0-2 double bonds). PIPn are precursors for second messengers. Polyunsaturated phosphatidylinositol bisphosphate (PIP2) exclusively supported signaling immediately upon T cell antigen receptor activation. In late Teff cells, activity of phospholipase C-γ1, the enzyme that cleaves PIP2 into downstream mediators, waned, and saturated PIPn became essential for sustained signaling. Saturated PIP was more rapidly converted to PIP2 with subsequent recruitment of phospholipase C-γ1, and loss of saturated PIPn impaired Teff cell fitness and function, even in cells with abundant polyunsaturated PIPn. Glucose was the substrate for de novo PIPn synthesis, and was rapidly utilized for saturated PIP2 generation. Thus, separate PIPn pools with distinct acyl chain compositions and metabolic dependencies drive important signaling events to initiate and then sustain effector function during CD8+ T cell differentiation.
© 2023. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests
E.L.P. is an SAB member of ImmunoMet Therapeutics and E.L.P. and E.J.P are Founders and Scientific Advisors to Rheos Medicines. The other authors declare no competing interests.
Figures


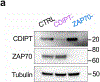








References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

