A palmitate-rich metastatic niche enables metastasis growth via p65 acetylation resulting in pro-metastatic NF-κB signaling
- PMID: 36732635
- PMCID: PMC7615234
- DOI: 10.1038/s43018-023-00513-2
A palmitate-rich metastatic niche enables metastasis growth via p65 acetylation resulting in pro-metastatic NF-κB signaling
Abstract
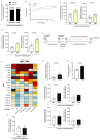
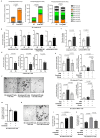
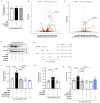
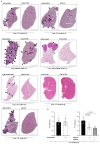
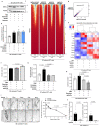
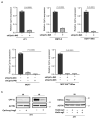
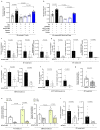
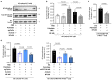
Metabolic rewiring is often considered an adaptive pressure limiting metastasis formation; however, some nutrients available at distant organs may inherently promote metastatic growth. We find that the lung and liver are lipid-rich environments. Moreover, we observe that pre-metastatic niche formation increases palmitate availability only in the lung, whereas a high-fat diet increases it in both organs. In line with this, targeting palmitate processing inhibits breast cancer-derived lung metastasis formation. Mechanistically, breast cancer cells use palmitate to synthesize acetyl-CoA in a carnitine palmitoyltransferase 1a-dependent manner. Concomitantly, lysine acetyltransferase 2a expression is promoted by palmitate, linking the available acetyl-CoA to the acetylation of the nuclear factor-kappaB subunit p65. Deletion of lysine acetyltransferase 2a or carnitine palmitoyltransferase 1a reduces metastasis formation in lean and high-fat diet mice, and lung and liver metastases from patients with breast cancer show coexpression of both proteins. In conclusion, palmitate-rich environments foster metastases growth by increasing p65 acetylation, resulting in a pro-metastatic nuclear factor-kappaB signaling.
© 2023. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
S-MF has received funding from Gilead, Bayer AG, Merck, Black Belt Therapeutics and Alesta Therapeutics, has consulted for Fund+ and is in the advisory board of Alesta Therapeutics. TGPG has consulted for Boehringer Ingelheim. All other authors declare no competing interests.
Figures









Comment in
-
Fatty acids prime the lung as a site for tumour spread.Nature. 2023 Mar;615(7951):224-225. doi: 10.1038/d41586-023-00538-8. Nature. 2023. PMID: 36854731 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

