Metabolomics profiling in prediction of chemo-immunotherapy efficiency in advanced non-small cell lung cancer
- PMID: 36733356
- PMCID: PMC9887290
- DOI: 10.3389/fonc.2022.1025046
Metabolomics profiling in prediction of chemo-immunotherapy efficiency in advanced non-small cell lung cancer
Abstract
Background: To explore potential metabolomics biomarker in predicting the efficiency of the chemo-immunotherapy in patients with advanced non-small cell lung cancer (NSCLC).
Methods: A total of 83 eligible patients were assigned to receive chemo-immunotherapy. Serum samples were prospectively collected before the treatment to perform metabolomics profiling analyses under the application of gas chromatography mass spectrometry (GC-MS). The key metabolites were identified using projection to latent structures discriminant analysis (PLS-DA). The key metabolites were used for predicting the chemo-immunotherapy efficiency in advanced NSCLC patients.
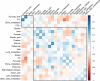
Results: Seven metabolites including pyruvate, threonine, alanine, urea, oxalate, elaidic acid and glutamate were identified as the key metabolites to the chemo-immunotherapy response. The receiver operating characteristic curves (AUC) were 0.79 (95% CI: 0.69-0.90), 0.60 (95% CI: 0.48-0.73), 0.69 (95% CI: 0.57-0.80), 0.63 (95% CI: 0.51-0.75), 0.60 (95% CI: 0.48-0.72), 0.56 (95% CI: 0.43-0.67), and 0.67 (95% CI: 0.55-0.80) for the key metabolites, respectively. A binary logistic regression was used to construct a combined biomarker model to improve the discriminating efficiency. The AUC was 0.86 (95% CI: 0.77-0.94) for the combined biomarker model. Pathway analyses showed that urea cycle, glucose-alanine cycle, glycine and serine metabolism, alanine metabolism, and glutamate metabolism were the key metabolic pathway to the chemo-immunotherapy response in patients with advanced NSCLC.
Conclusion: Metabolomics analyses of key metabolites and pathways revealed that GC-MS could be used to predict the efficiency of chemo-immunotherapy. Pyruvate, threonine, alanine, urea, oxalate, elaidic acid and glutamate played a central role in the metabolic of PD patients with advanced NSCLC.
Keywords: GC-MS; biomarker; chemo-immunotherapy; metabolomics; non-small cell lung cancer.
Copyright © 2023 Mei, Zhang, Li, Yang and Qi.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Xiang C, Han Y, Teng H, Zhu L, Lizaso A. Comprehensive investigation of mutational features of various lung adenocarcinoma histological subtypes among chinese patients. J Thorac Oncol (2021) 16:S460. doi: 10.1016/j.jtho.2021.01.788 - DOI
-
- Gharibvand L, Beeson WL, Shavlik D, Knutsen R, Knutsen SF. Lung adenocarcinoma and ambient particulate air pollution. Ann Epidemiol (2017) 27:506–7. doi: 10.1016/j.annepidem.2017.07.116 - DOI
-
- Paz-Ares L, Ciuleanu TE, Cobo M, Schenker M, Zurawski B, Menezes J, et al. First-line nivolumab plus ipilimumab combined with two cycles of chemotherapy in patients with non-small-cell lung cancer (CheckMate 9LA): an international, randomised, open-label, phase 3 trial. Lancet Oncol (2021) 22:198–211. doi: 10.1016/S1470-2045(20)30641-0 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

