Comparative transcriptome analysis of rainbow trout gonadal cells (RTG-2) infected with U and J genogroup infectious hematopoietic necrosis virus
- PMID: 36733771
- PMCID: PMC9887044
- DOI: 10.3389/fmicb.2022.1109606
Comparative transcriptome analysis of rainbow trout gonadal cells (RTG-2) infected with U and J genogroup infectious hematopoietic necrosis virus
Abstract
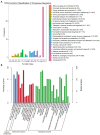
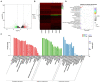
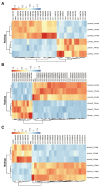
Infectious hematopoietic necrosis virus (IHNV) is the causative pathogen of infectious hematopoietic necrosis, outbreaks of which are responsible for significant losses in rainbow trout aquaculture. Strains of IHNV isolated worldwide have been classified into five major genogroups, J, E, L, M, and U. To date, comparative transcriptomic analysis has only been conducted individually for the J and M genogroups. In this study, we compared the transcriptome profiles in U genogroup and J genogroup IHNV-infected RTG-2 cells with mock-infected RTG-2 cells. The RNA-seq results revealed 17,064 new genes, of which 7,390 genes were functionally annotated. Differentially expressed gene (DEG) analysis between U and J IHNV-infected cells revealed 2,238 DEGs, including 1,011 downregulated genes and 1,227 upregulated genes. Among the 2,238 DEGs, 345 new genes were discovered. The DEGs related to immune responses, cellular signal transduction, and viral diseases were further analyzed. RT-qPCR validation confirmed that the changes in expression of the immune response-related genes trpm2, sting, itgb7, ripk2, and irf1, cellular signal transduction-related genes irl, cacnb2, bmp2l, gadd45α, and plk2, and viral disease-related genes mlf1, mtor, armc5, pik3r1, and c-myc were consistent with the results of transcriptome analysis. Taken together, our findings provide a comprehensive transcriptional analysis of the differential virulence of the U and J genogroups of IHNV, and shed new light on the pathogenic mechanisms of IHNV strains.
Keywords: J genogroup; U genogroup; infectious hematopoietic necrosis virus; rainbow trout gonadal cells; transcriptome analysis (RNAseq).
Copyright © 2023 Zhao, Xu, Ren, Shao, Liu, Teng and Lu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

