Generative power of a protein language model trained on multiple sequence alignments
- PMID: 36734516
- PMCID: PMC10038667
- DOI: 10.7554/eLife.79854
Generative power of a protein language model trained on multiple sequence alignments
Abstract
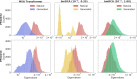
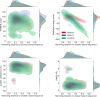
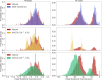
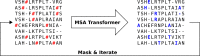
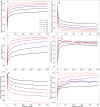
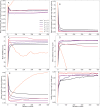
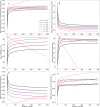
Computational models starting from large ensembles of evolutionarily related protein sequences capture a representation of protein families and learn constraints associated to protein structure and function. They thus open the possibility for generating novel sequences belonging to protein families. Protein language models trained on multiple sequence alignments, such as MSA Transformer, are highly attractive candidates to this end. We propose and test an iterative method that directly employs the masked language modeling objective to generate sequences using MSA Transformer. We demonstrate that the resulting sequences score as well as natural sequences, for homology, coevolution, and structure-based measures. For large protein families, our synthetic sequences have similar or better properties compared to sequences generated by Potts models, including experimentally validated ones. Moreover, for small protein families, our generation method based on MSA Transformer outperforms Potts models. Our method also more accurately reproduces the higher-order statistics and the distribution of sequences in sequence space of natural data than Potts models. MSA Transformer is thus a strong candidate for protein sequence generation and protein design.
Keywords: computational biology; deep learning; none; protein design; protein families; protein language models; protein sequence generation; protein sequences; systems biology.
© 2023, Sgarbossa et al.
Conflict of interest statement
DS, UL, AB No competing interests declared
Figures
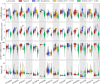

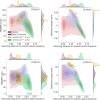
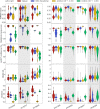

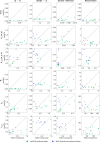


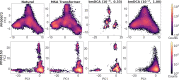
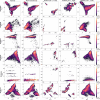
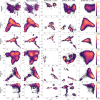
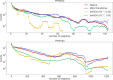


Update of
- doi: 10.1101/2022.04.14.488405
Similar articles
-
Protein language models trained on multiple sequence alignments learn phylogenetic relationships.Nat Commun. 2022 Oct 22;13(1):6298. doi: 10.1038/s41467-022-34032-y. Nat Commun. 2022. PMID: 36273003 Free PMC article.
-
Pairing interacting protein sequences using masked language modeling.Proc Natl Acad Sci U S A. 2024 Jul 2;121(27):e2311887121. doi: 10.1073/pnas.2311887121. Epub 2024 Jun 24. Proc Natl Acad Sci U S A. 2024. PMID: 38913900 Free PMC article.
-
Phylogenetic Corrections and Higher-Order Sequence Statistics in Protein Families: The Potts Model vs MSA Transformer.ArXiv [Preprint]. 2025 Mar 1:arXiv:2503.00289v1. ArXiv. 2025. PMID: 40365615 Free PMC article. Preprint.
-
Recent advances in features generation for membrane protein sequences: From multiple sequence alignment to pre-trained language models.Proteomics. 2023 Dec;23(23-24):e2200494. doi: 10.1002/pmic.202200494. Epub 2023 Oct 20. Proteomics. 2023. PMID: 37863817 Review.
-
Transformer-based deep learning for predicting protein properties in the life sciences.Elife. 2023 Jan 18;12:e82819. doi: 10.7554/eLife.82819. Elife. 2023. PMID: 36651724 Free PMC article. Review.
Cited by
-
Protein language models trained on multiple sequence alignments learn phylogenetic relationships.Nat Commun. 2022 Oct 22;13(1):6298. doi: 10.1038/s41467-022-34032-y. Nat Commun. 2022. PMID: 36273003 Free PMC article.
-
Pairing interacting protein sequences using masked language modeling.Proc Natl Acad Sci U S A. 2024 Jul 2;121(27):e2311887121. doi: 10.1073/pnas.2311887121. Epub 2024 Jun 24. Proc Natl Acad Sci U S A. 2024. PMID: 38913900 Free PMC article.
-
Direct coupling analysis and the attention mechanism.BMC Bioinformatics. 2025 Feb 6;26(1):41. doi: 10.1186/s12859-025-06062-y. BMC Bioinformatics. 2025. PMID: 39915710 Free PMC article.
-
Generative Artificial Intelligence-Assisted Protein Design Must Consider Repurposing Potential.GEN Biotechnol. 2023 Aug 1;2(4):296-300. doi: 10.1089/genbio.2023.0025. Epub 2023 Aug 17. GEN Biotechnol. 2023. PMID: 37928405 Free PMC article.
-
Opportunities and Challenges for Machine Learning-Assisted Enzyme Engineering.ACS Cent Sci. 2024 Feb 5;10(2):226-241. doi: 10.1021/acscentsci.3c01275. eCollection 2024 Feb 28. ACS Cent Sci. 2024. PMID: 38435522 Free PMC article. Review.
References
-
- Baek M, DiMaio F, Anishchenko I, Dauparas J, Ovchinnikov S, Lee GR, Wang J, Cong Q, Kinch LN, Schaeffer RD, Millán C, Park H, Adams C, Glassman CR, DeGiovanni A, Pereira JH, Rodrigues AV, van Dijk AA, Ebrecht AC, Opperman DJ, Sagmeister T, Buhlheller C, Pavkov-Keller T, Rathinaswamy MK, Dalwadi U, Yip CK, Burke JE, Garcia KC, Grishin NV, Adams PD, Read RJ, Baker D. Accurate prediction of protein structures and interactions using a three-track neural network. Science. 2021;373:871–876. doi: 10.1126/science.abj8754. - DOI - PMC - PubMed
-
- Bahdanau D, Cho K, Bengio Y. Neural machine translation by jointly learning to align and translate. International Conference on Learning Representations.2015.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

