Cytidine deaminases catalyze the conversion of N(S, O)4-substituted pyrimidine nucleosides
- PMID: 36735785
- PMCID: PMC9897663
- DOI: 10.1126/sciadv.ade4361
Cytidine deaminases catalyze the conversion of N(S, O)4-substituted pyrimidine nucleosides
Abstract
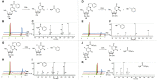
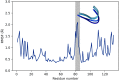
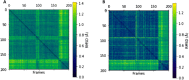
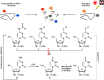
Cytidine deaminases (CDAs) catalyze the hydrolytic deamination of cytidine and 2'-deoxycytidine to uridine and 2'-deoxyuridine. Here, we report that prokaryotic homo-tetrameric CDAs catalyze the nucleophilic substitution at the fourth position of N4-acyl-cytidines, N4-alkyl-cytidines, and N4-alkyloxycarbonyl-cytidines, and S4-alkylthio-uridines and O4-alkyl-uridines, converting them to uridine and corresponding amide, amine, carbamate, thiol, or alcohol as leaving groups. The x-ray structure of a metagenomic CDA_F14 and the molecular modeling of the CDAs used in this study show a relationship between the bulkiness of a leaving group and the volume of the binding pocket, which is partly determined by the flexible β3α3 loop of CDAs. We propose that CDAs that are active toward a wide range of substrates participate in salvage and/or catabolism of variously modified pyrimidine nucleosides. This identified promiscuity of CDAs expands the knowledge about the cellular turnover of cytidine derivatives, including the pharmacokinetics of pyrimidine-based prodrugs.
Figures


References
-
- P. Boccaletto, F. Stefaniak, A. Ray, A. Cappannini, S. Mukherjee, E. Purta, M. Kurkowska, N. Shirvanizadeh, E. Destefanis, P. Groza, G. Avşar, A. Romitelli, P. Pir, E. Dassi, S. G. Conticello, F. Aguilo, J. M. Bujnicki, MODOMICS: A database of RNA modification pathways. 2021 update. Nucleic Acids Res. 50, D231–D235 (2022). - PMC - PubMed
-
- W. L. Nyhan, Nucleotide Synthesis Via Salvage Pathway, in eLS (John Wiley & Sons Ltd, 2014).
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

