Alternative polyadenylation transcriptome-wide association study identifies APA-linked susceptibility genes in brain disorders
- PMID: 36737438
- PMCID: PMC9898543
- DOI: 10.1038/s41467-023-36311-8
Alternative polyadenylation transcriptome-wide association study identifies APA-linked susceptibility genes in brain disorders
Abstract
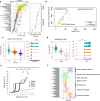
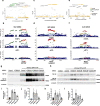
Alternative polyadenylation (APA) plays an essential role in brain development; however, current transcriptome-wide association studies (TWAS) largely overlook APA in nominating susceptibility genes. Here, we performed a 3' untranslated region (3'UTR) APA TWAS (3'aTWAS) for 11 brain disorders by combining their genome-wide association studies data with 17,300 RNA-seq samples across 2,937 individuals. We identified 354 3'aTWAS-significant genes, including known APA-linked risk genes, such as SNCA in Parkinson's disease. Among these 354 genes, ~57% are not significant in traditional expression- and splicing-TWAS studies, since APA may regulate the translation, localization and protein-protein interaction of the target genes independent of mRNA level expression or splicing. Furthermore, we discovered ATXN3 as a 3'aTWAS-significant gene for amyotrophic lateral sclerosis, and its modulation substantially impacted pathological hallmarks of amyotrophic lateral sclerosis in vitro. Together, 3'aTWAS is a powerful strategy to nominate important APA-linked brain disorder susceptibility genes, most of which are largely overlooked by conventional expression and splicing analyses.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

