Human-specific genetics: new tools to explore the molecular and cellular basis of human evolution
- PMID: 36737647
- PMCID: PMC9897628
- DOI: 10.1038/s41576-022-00568-4
Human-specific genetics: new tools to explore the molecular and cellular basis of human evolution
Abstract
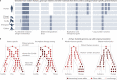
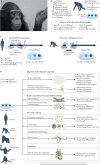
Our ancestors acquired morphological, cognitive and metabolic modifications that enabled humans to colonize diverse habitats, develop extraordinary technologies and reshape the biosphere. Understanding the genetic, developmental and molecular bases for these changes will provide insights into how we became human. Connecting human-specific genetic changes to species differences has been challenging owing to an abundance of low-effect size genetic changes, limited descriptions of phenotypic differences across development at the level of cell types and lack of experimental models. Emerging approaches for single-cell sequencing, genetic manipulation and stem cell culture now support descriptive and functional studies in defined cell types with a human or ape genetic background. In this Review, we describe how the sequencing of genomes from modern and archaic hominins, great apes and other primates is revealing human-specific genetic changes and how new molecular and cellular approaches - including cell atlases and organoids - are enabling exploration of the candidate causal factors that underlie human-specific traits.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

