Gut microbiome variations in Rhinopithecus roxellanae caused by changes in the environment
- PMID: 36737703
- PMCID: PMC9896789
- DOI: 10.1186/s12864-023-09142-6
Gut microbiome variations in Rhinopithecus roxellanae caused by changes in the environment
Abstract
Background: The snub-nosed monkey (Rhinopithecus roxellanae) is an endangered animal species mainly distributed in China and needs to be protected. Gut microbiome is an important determinant of animal health and population survival as it affects the adaptation of the animals to different foods and environments under kinetic changes of intrinsic and extrinsic factors. Therefore, this study aimed to elucidate gut fecal microbiome profiles of snub-nosed monkeys affected by several extrinsic and intrinsic factors, including raising patterns (captive vs. wild), age, sex, and diarrheal status to provide a reference for making protection strategies.
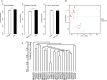
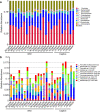
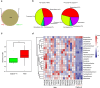
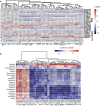
Results: The 16S rRNA gene sequencing was firstly used to pre-check clustering of 38 fecal samples from the monkeys including 30 wild and 8 captive (5 healthy and 3 diarrheal) from three Regions of Shennongjia Nature Reserve, Hubei Province, China. Then the 24 samples with high-quality DNA from 18 wild and 6 captive (4 healthy and 2 diarrheal) monkeys were subjected to shotgun metagenomic sequencing to characterize bacterial gut microbial communities. We discovered that the raising pattern (captive and wild) rather than age and sex was the predominant factor attributed to gut microbiome structure and proportionality. Wild monkeys had significantly higher bacterial diversity and lower Bacteroidetes/Firmicutes ratios than captive animals. Moreover, the gut microbiomes in wild healthy monkeys were enriched for the genes involved in fatty acid production, while in captive animals, genes were enriched for vitamin biosynthesis and metabolism and amino acid biosynthesis from carbohydrate intermediates. Additionally, a total of 37 antibiotic resistant genes (ARG) types were detected. Unlike the microbiome diversity, the captive monkeys have a higher diversity of ARG than the wild animals.
Conclusion: Taken together, we highlight the importance of self-reprogramed metabolism in the snub-nosed monkey gut microbiome to help captive and wild monkeys adapt to different intrinsic and extrinsic environmental change.
Keywords: Adaptation; Antibiotic resistant genes; Bacteroidetes/Firmicutes ratio; Gut microbiome; Metagenomics; Rhinopithecus roxellanae.
© 2023. The Author(s).
Conflict of interest statement
We declare that we have no competing interests.
Figures


References
-
- Long YR, Richardson M. Rhinopithecus roxellana. The IUCN Red List of Threatened Species 2020:e.T1959A17943886. 10.2305/IUCN.UK2020-2.RLTS.T1959A17943886.en. - DOI
-
- Luo M, Liu Z, Pan H, et al. Historical geographic dispersal of the golden snub-nosed monkey (Rhinopithecus roxellana) and the influence of climatic oscillations. Am J Primatol. 2012;74:91–101. - PubMed
-
- Zhou X, Meng X, Liu Z, et al. Population genomics reveals low genetic diversity and adaptation to hypoxia in snub-nosed monkeys. Mol Biol Evol. 2016;33:2670–2681. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

