Genome-wide screening reveals the genetic basis of mammalian embryonic eye development
- PMID: 36737727
- PMCID: PMC9898963
- DOI: 10.1186/s12915-022-01475-0
Genome-wide screening reveals the genetic basis of mammalian embryonic eye development
Abstract
Background: Microphthalmia, anophthalmia, and coloboma (MAC) spectrum disease encompasses a group of eye malformations which play a role in childhood visual impairment. Although the predominant cause of eye malformations is known to be heritable in nature, with 80% of cases displaying loss-of-function mutations in the ocular developmental genes OTX2 or SOX2, the genetic abnormalities underlying the remaining cases of MAC are incompletely understood. This study intended to identify the novel genes and pathways required for early eye development. Additionally, pathways involved in eye formation during embryogenesis are also incompletely understood. This study aims to identify the novel genes and pathways required for early eye development through systematic forward screening of the mammalian genome.
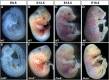
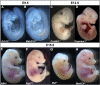
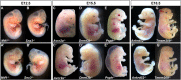
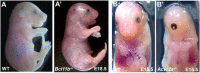
Results: Query of the International Mouse Phenotyping Consortium (IMPC) database (data release 17.0, August 01, 2022) identified 74 unique knockout lines (genes) with genetically associated eye defects in mouse embryos. The vast majority of eye abnormalities were small or absent eyes, findings most relevant to MAC spectrum disease in humans. A literature search showed that 27 of the 74 lines had previously published knockout mouse models, of which only 15 had ocular defects identified in the original publications. These 12 previously published gene knockouts with no reported ocular abnormalities and the 47 unpublished knockouts with ocular abnormalities identified by the IMPC represent 59 genes not previously associated with early eye development in mice. Of these 59, we identified 19 genes with a reported human eye phenotype. Overall, mining of the IMPC data yielded 40 previously unimplicated genes linked to mammalian eye development. Bioinformatic analysis showed that several of the IMPC genes colocalized to several protein anabolic and pluripotency pathways in early eye development. Of note, our analysis suggests that the serine-glycine pathway producing glycine, a mitochondrial one-carbon donator to folate one-carbon metabolism (FOCM), is essential for eye formation.
Conclusions: Using genome-wide phenotype screening of single-gene knockout mouse lines, STRING analysis, and bioinformatic methods, this study identified genes heretofore unassociated with MAC phenotypes providing models to research novel molecular and cellular mechanisms involved in eye development. These findings have the potential to hasten the diagnosis and treatment of this congenital blinding disease.
Keywords: CPLANE; Eye development; IMPC; MAC spectrum; Mouse; Serine-glycine biosynthesis.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
-
- Graw J. Eye development. Curr Top Dev Biol. 2010;90:343–386. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

