A human liver organoid screening platform for DILI risk prediction
- PMID: 36738840
- PMCID: PMC11268729
- DOI: 10.1016/j.jhep.2023.01.019
A human liver organoid screening platform for DILI risk prediction
Abstract
Background & aims: Drug-induced liver injury (DILI), both intrinsic and idiosyncratic, causes frequent morbidity, mortality, clinical trial failures and post-approval withdrawal. This suggests an unmet need for improved in vitro models for DILI risk prediction that can account for diverse host genetics and other clinical factors. In this study, we evaluated the utility of human liver organoids (HLOs) for high-throughput DILI risk prediction and in an organ-on-chip system.
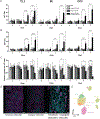
Methods: HLOs were derived from three separate iPSC lines and benchmarked on two platforms for their ability to model in vitro liver function and identify hepatotoxic compounds using biochemical assays for albumin, ALT, AST, microscopy-based morphological profiling, and single-cell transcriptomics: i) HLOs dispersed in 384-well-formatted plates and exposed to a library of compounds; ii) HLOs adapted to a liver-on-chip system.
Results: Dispersed HLOs derived from the three iPSC lines had similar DILI predictive capacity as intact HLOs in a high-throughput screening format, allowing for measurable IC50 values of compound cytotoxicity. Distinct morphological differences were observed in cells treated with drugs exerting differing mechanisms of toxicity. On-chip HLOs significantly increased albumin production, CYP450 expression, and ALT/AST release when treated with known hepatoxic drugs compared to dispersed HLOs and primary human hepatocytes. On-chip HLOs were able to predict the synergistic hepatotoxicity of tenofovir-inarigivir and displayed steatosis and mitochondrial perturbation, via phenotypic and transcriptomic analysis, on exposure to fialuridine and acetaminophen, respectively.
Conclusions: The high-throughput and liver-on-chip systems exhibit enhanced in vivo-like functions and demonstrate the potential utility of these platforms for DILI risk assessment. Tenofovir-inarigivr-associated hepatotoxicity was observed and correlates with the clinical manifestation of DILI observed in patients.
Impact and implications: Idiosyncratic (spontaneous, patient-specific) drug-induced liver injury (DILI) is difficult to study due to the lack of liver models that function as human liver tissue and are adaptable for large-scale drug screening. Human liver organoids grown from patient stem cells respond to known DILI-causing drugs in both a high-throughput and on a physiological "chip" culture system. These platforms show promise for researchers in their use as predictive models for novel drugs before entering clinical trials and as a potential in vitro diagnostic tool. Our findings support further development of patient-derived liver organoid lines and their use in the context of DILI research.
Keywords: drug development; hepatotoxicity; high-content imaging; microfluidic devices.
Copyright © 2023 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Conflicts of interest: RJF has research support from Gilead and Abbvie. Other authors have no conflicts.
Figures
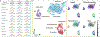


References
-
- Bakke OM, Manocchia M, de Abajo F, Kaitin KI, Lasagna L. Drug safety discontinuations in the United Kingdom, the United States, and Spain from 1974 through 1993: a regulatory perspective. Clin. Pharmacol. Ther. 1995;58:108–117. - PubMed
-
- Watkins PB. Drug safety sciences and the bottleneck in drug development. Clin. Pharmacol. Ther. 2011;89:788–790. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

