Vast heterogeneity in cytoplasmic diffusion rates revealed by nanorheology and Doppelgänger simulations
- PMID: 36739478
- PMCID: PMC10027447
- DOI: 10.1016/j.bpj.2023.01.040
Vast heterogeneity in cytoplasmic diffusion rates revealed by nanorheology and Doppelgänger simulations
Abstract
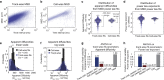
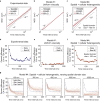
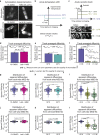
The cytoplasm is a complex, crowded, actively driven environment whose biophysical characteristics modulate critical cellular processes such as cytoskeletal dynamics, phase separation, and stem cell fate. Little is known about the variance in these cytoplasmic properties. Here, we employed particle-tracking nanorheology on genetically encoded multimeric 40 nm nanoparticles (GEMs) to measure diffusion within the cytoplasm of individual fission yeast (Schizosaccharomyces pombe) cellscells. We found that the apparent diffusion coefficients of individual GEM particles varied over a 400-fold range, while the differences in average particle diffusivity among individual cells spanned a 10-fold range. To determine the origin of this heterogeneity, we developed a Doppelgänger simulation approach that uses stochastic simulations of GEM diffusion that replicate the experimental statistics on a particle-by-particle basis, such that each experimental track and cell had a one-to-one correspondence with their simulated counterpart. These simulations showed that the large intra- and inter-cellular variations in diffusivity could not be explained by experimental variability but could only be reproduced with stochastic models that assume a wide intra- and inter-cellular variation in cytoplasmic viscosity. The simulation combining intra- and inter-cellular variation in viscosity also predicted weak nonergodicity in GEM diffusion, consistent with the experimental data. To probe the origin of this variation, we found that the variance in GEM diffusivity was largely independent of factors such as temperature, the actin and microtubule cytoskeletons, cell-cyle stage, and spatial locations, but was magnified by hyperosmotic shocks. Taken together, our results provide a striking demonstration that the cytoplasm is not "well-mixed" but represents a highly heterogeneous environment in which subcellular components at the 40 nm size scale experience dramatically different effective viscosities within an individual cell, as well as in different cells in a genetically identical population. These findings carry significant implications for the origins and regulation of biological noise at cellular and subcellular levels.
Copyright © 2023 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



References
-
- Schmoller K.M. The phenomenology of cell size control. Curr. Opin. Cell Biol. 2017;49:53–58. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

