Disordered proteins mitigate the temperature dependence of site-specific binding free energies
- PMID: 36739945
- PMCID: PMC10027511
- DOI: 10.1016/j.jbc.2023.102984
Disordered proteins mitigate the temperature dependence of site-specific binding free energies
Abstract
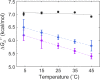
Biophysical characterization of protein-protein interactions involving disordered proteins is challenging. A common simplification is to measure the thermodynamics and kinetics of disordered site binding using peptides containing only the minimum residues necessary. We should not assume, however, that these few residues tell the whole story. Son of sevenless, a multidomain signaling protein from Drosophila melanogaster, is critical to the mitogen-activated protein kinase pathway, passing an external signal to Ras, which leads to cellular responses. The disordered 55 kDa C-terminal domain of Son of sevenless is an autoinhibitor that blocks guanidine exchange factor activity. Activation requires another protein, Downstream of receptor kinase (Drk), which contains two Src homology 3 domains. Here, we utilized NMR spectroscopy and isothermal titration calorimetry to quantify the thermodynamics and kinetics of the N-terminal Src homology 3 domain binding to the strongest sites incorporated into the flanking disordered sequences. Comparing these results to those for isolated peptides provides information about how the larger domain affects binding. The affinities of sites on the disordered domain are like those of the peptides at low temperatures but less sensitive to temperature. Our results, combined with observations showing that intrinsically disordered proteins become more compact with increasing temperature, suggest a mechanism for this effect.
Keywords: Src homology 3 domain; biophysics; intrinsically disordered protein; protein–protein interaction; thermodynamics.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare no conflicts of interests with the contents of this article.
Figures



References
-
- Xue B., Dunker A.K., Uversky V.N. Orderly order in protein intrinsic disorder distribution: disorder in 3500 proteomes from viruses and the three domains of life. J. Biomol. Struct. Dyn. 2012;30:137–149. - PubMed
-
- Dunker A.K., Lawson J.D., Brown C.J., Williams R.M., Romero P., Oh J.S., et al. Intrinsically disordered protein. J. Mol. Graphics Model. 2001;19:26–59. - PubMed
-
- Wright P.E., Dyson H.J. Intrinsically unstructured proteins: Re-assessing the protein structure-function paradigm. J. Mol. Biol. 1999;293:321–331. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

