Immersion Fixation and Staining of Multicubic Millimeter Volumes for Electron Microscopy-Based Connectomics of Human Brain Biopsies
- PMID: 36740206
- PMCID: PMC10397365
- DOI: 10.1016/j.biopsych.2023.01.025
Immersion Fixation and Staining of Multicubic Millimeter Volumes for Electron Microscopy-Based Connectomics of Human Brain Biopsies
Abstract
Connectomics allows mapping of cells and their circuits at the nanometer scale in volumes of approximately 1 mm3. Given that the human cerebral cortex can be 3 mm in thickness, larger volumes are required. Larger-volume circuit reconstructions of human brain are limited by 1) the availability of fresh biopsies; 2) the need for excellent preservation of ultrastructure, including extracellular space; and 3) the requirement of uniform staining throughout the sample, among other technical challenges. Cerebral cortical samples from neurosurgical patients are available owing to lead placement for deep brain stimulation. Described here is an immersion fixation, heavy metal staining, and tissue processing method that consistently provides excellent ultrastructure throughout human and rodent surgical brain samples of volumes 2 × 2 × 2 mm3 and up to 37 mm3 with one dimension ≤2 mm. This method should allow synapse-level circuit analysis in samples from patients with psychiatric and neurologic disorders.
Keywords: Brain biopsy; Connectomics; Electron microscopy; Human cortex; Immersion fixation; Osmium staining.
Copyright © 2023 Society of Biological Psychiatry. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Disclosure
The authors declare no biomedical financial interests nor conflict of interest.
Figures
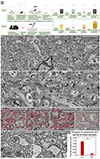
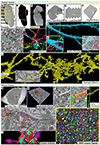
Comment in
-
Looking at the Human Brain in Detail.Biol Psychiatry. 2023 Aug 15;94(4):285-287. doi: 10.1016/j.biopsych.2023.06.006. Biol Psychiatry. 2023. PMID: 37495332 No abstract available.
References
-
- Kasthuri N, Hayworth KJ, Berger DR, Schalek RL, Conchello JA, Knowles-Barley S, et al. (2015): Saturated reconstruction of a volume of neocortex. Cell 162: 648–661. - PubMed
-
- Mikula S, Denk W (2015): High-resolution whole-brain staining for electron microscopic circuit reconstruction. Nat Methods 12: 541–546. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

