The epigenetic landscape of oligodendrocyte lineage cells
- PMID: 36740586
- PMCID: PMC10085863
- DOI: 10.1111/nyas.14959
The epigenetic landscape of oligodendrocyte lineage cells
Abstract
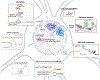
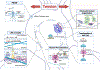
The epigenetic landscape of oligodendrocyte lineage cells refers to the cell-specific modifications of DNA, chromatin, and RNA that define a unique gene expression pattern of functionally specialized cells. Here, we focus on the epigenetic changes occurring as progenitors differentiate into myelin-forming cells and respond to the local environment. First, modifications of DNA, RNA, nucleosomal histones, key principles of chromatin organization, topologically associating domains, and local remodeling will be reviewed. Then, the relationship between epigenetic modulators and RNA processing will be explored. Finally, the reciprocal relationship between the epigenome as a determinant of the mechanical properties of cell nuclei and the target of mechanotransduction will be discussed. The overall goal is to provide an interpretative key on how epigenetic changes may account for the heterogeneity of the transcriptional profiles identified in this lineage.
Keywords: brain; chromatin; development; histone; myelin; transcription.
© 2023 New York Academy of Sciences.
Conflict of interest statement
COMPETING INTERESTS
The authors declare no competing interests.
Figures

References
-
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, & van Steensel B (2008). Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature, 453, 948–951. - PubMed
-
- Towbin BD, González-Aguilera C, Sack R, Gaidatzis D, Kalck V, Meister P, Askjaer P, & Gasser SM (2012). Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell, 150, 934–947. - PubMed
-
- Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, & Cavalli G (2012). Three-dimensional folding and functional organization principles of the Drosophila genome. Cell, 148, 458–472. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

