Diffusion time dependency of extracellular diffusion
- PMID: 36740894
- PMCID: PMC10392121
- DOI: 10.1002/mrm.29594
Diffusion time dependency of extracellular diffusion
Abstract
Purpose: To quantify the variations of the power-law dependences on diffusion time t or gradient frequency of extracellular water diffusion measured by diffusion MRI (dMRI).
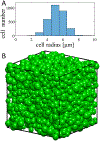
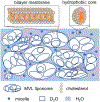
Methods: Model cellular systems containing only extracellular water were used to investigate the dependence of , the extracellular diffusion coefficient. Computer simulations used a randomly packed tissue model with realistic intracellular volume fractions and cell sizes. DMRI measurements were performed on samples consisting of liposomes containing heavy water(D2 O, deuterium oxide) dispersed in regular water (H2 O). was obtained over a broad range (∼1-1000 ms) and then fit power-law equations and .
Results: Both simulated and experimental results suggest that no single power-law adequately describes the behavior of over the range of diffusion times of most interest in practical dMRI. Previous theoretical predictions are accurate over only limited ranges; for example, is valid only for short times, whereas or is valid only for long times but cannot describe other ranges simultaneously. For the specific range of 5-70 ms used in typical human dMRI measurements, matches the data well empirically.
Conclusion: The optimal power-law fit of extracellular diffusion varies with diffusion time. The dependency obtained at short or long limits cannot be applied to typical dMRI measurements in human cancer or liver. It is essential to determine the appropriate diffusion time range when modeling extracellular diffusion in dMRI-based quantitative microstructural imaging.
Keywords: D2O; diffusion; diffusion time; extracellular; liposome; oscillating gradient; phantom.
© 2023 International Society for Magnetic Resonance in Medicine.
Figures



References
-
- Panagiotaki E, Walker-Samuel S, Siow B, Johnson SP, Rajkumar V, Pedley RB, Lythgoe MF, Alexander DC. Noninvasive quantification of solid tumor microstructure using VERDICT MRI. Cancer Res 2014;74(7):1902–1912. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

