Design of a chimeric ACE-2/Fc-silent fusion protein with ultrahigh affinity and neutralizing capacity for SARS-CoV-2 variants
- PMID: 36741194
- PMCID: PMC9889962
- DOI: 10.1093/abt/tbad001
Design of a chimeric ACE-2/Fc-silent fusion protein with ultrahigh affinity and neutralizing capacity for SARS-CoV-2 variants
Abstract
Background: As SARS-CoV-2 continues to mutate into Variants of Concern (VOC), there is growing and urgent need to develop effective antivirals to combat COVID-19. Monoclonal antibodies developed earlier are no longer capable of effectively neutralizing currently active VOCs. This report describes the design of variant-agnostic chimeric molecules consisting of an Angiotensin-Converting Enzyme 2 (ACE-2) domain mutated to retain ultrahigh affinity binding to a wide variety of SARS-CoV-2 variants, coupled to an Fc-silent immunoglobulin domain that eliminates antibody-dependent enhancement and extends biological half-life.
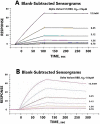
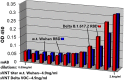
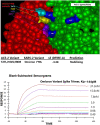
Methods: Molecular modeling, Surrogate Viral Neutralization tests (sVNTs) and infection studies of human airway organoid cultures were performed with synthetic chimeras, SARS-CoV-2 spike protein mimics and SARS-CoV-2 Omicron variants B.1.1.214, BA.1, BA.2 and BA.5.
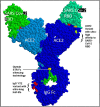
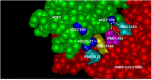
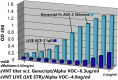
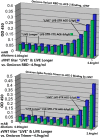
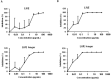
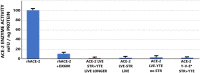
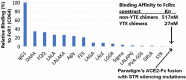
Results: ACE-2 mutations L27, V34 and E90 resulted in ultrahigh affinity binding of the LVE-ACE-2 domain to the widest variety of VOCs, with KDs of 93 pM and 73 pM for binding to the Alpha B1.1.7 and Omicron B.1.1.529 variants, and notably, 78fM, 133fM and 1.81pM affinities to the Omicron BA.2, BA2.75 and BQ.1.1 subvariants, respectively. sVNT assays revealed titers of ≥4.9 ng/ml, for neutralization of recombinant viral proteins corresponding to the Alpha, Delta and Omicron variants. The values above were obtained with LVE-ACE-2/mAB chimeras containing the FcRn-binding Y-T-E sequence which extends biological half-life 3-4-fold.
Conclusions: The ACE-2-mutant/Fc silent fusion proteins described have ultrahigh affinity to a wide variety of SARS-CoV-2 variants including Omicron. It is proposed that these chimeric ACE-2/mABs will constitute variant-agnostic and cost-effective prophylactics against SARS-CoV-2, particularly when administered nasally.
Keywords: ACE-2; coronavirus; fusion protein; lung; therapeutic.
© The Author(s) 2023. Published by Oxford University Press on behalf of Antibody Therapeutics. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures






References
-
- Heurich, A, Hofmann-Winkler, H, Gierer, Set al. TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. J Virol 2014; 88: 1293–307Epub 2013 Nov 13. PMID: 24227843; PMCID: PMC3911672. - PMC - PubMed
-
- Celik, I, Khan, A, Dwivany, FMet al. Computational prediction of the effect of mutations in the receptor-binding domain on the interaction between SARS-CoV-2 and human ACE2. Mol Divers 2022; 26: 3309–24PMID: 35138508. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
