Blood transcriptome responses in patients correlate with severity of COVID-19 disease
- PMID: 36741372
- PMCID: PMC9896980
- DOI: 10.3389/fimmu.2022.1043219
Blood transcriptome responses in patients correlate with severity of COVID-19 disease
Abstract
Background: Coronavirus disease 2019 (COVID-19) is an infectious disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Infected individuals display a wide spectrum of disease severity, as defined by the World Health Organization (WHO). One of the main factors underlying this heterogeneity is the host immune response, with severe COVID-19 often associated with a hyperinflammatory state.
Aim: Our current study aimed to pinpoint the specific genes and pathways underlying differences in the disease spectrum and outcomes observed, through in-depth analyses of whole blood transcriptomics in a large cohort of COVID-19 participants.
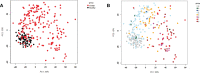
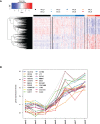
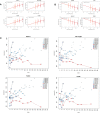
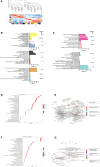
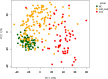
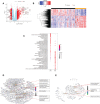
Results: All WHO severity levels were well represented and mild and severe disease displaying distinct gene expression profiles. WHO severity levels 1-4 were grouped as mild disease, and signatures from these participants were different from those with WHO severity levels 6-9 classified as severe disease. Severity level 5 (moderate cases) presented a unique transitional gene signature between severity levels 2-4 (mild/moderate) and 6-9 (severe) and hence might represent the turning point for better or worse disease outcome. Gene expression changes are very distinct when comparing mild/moderate or severe cases to healthy controls. In particular, we demonstrated the hallmark down-regulation of adaptive immune response pathways and activation of neutrophil pathways in severe compared to mild/moderate cases, as well as activation of blood coagulation pathways.
Conclusions: Our data revealed discrete gene signatures associated with mild, moderate, and severe COVID-19 identifying valuable candidates for future biomarker discovery.
Keywords: RNA sequencing; SARS-CoV-2; WGCNA; deconvolution; host immune response.
Copyright © 2023 Wang, Schughart, Pelaia, Chew, Kim, Karvunidis, Knippenberg, Teoh, Phu, Short, Iredell, Thevarajan, Audsley, Macdonald, Burcham, McLean, PREDICT-19 consortium, Tang and Shojaei.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

