Unravelling the fungal darkness in a tropical cave: richness and the description of one new genus and six new species
- PMID: 36741552
- PMCID: PMC9875697
- DOI: 10.3114/fuse.2022.10.06
Unravelling the fungal darkness in a tropical cave: richness and the description of one new genus and six new species
Abstract
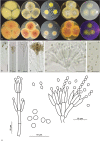
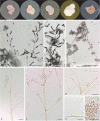
Caves are special environments that harbour an incredible diversity of life, including fungal species. Brazilian caves have been demonstrated to be biodiversity hotspots for known and unknown fungal species. We investigated the richness of culturable fungi in a tropical cave in Brazil by isolating these microorganisms from the sediment and air. The fungal abundance of colony-forming units (CFUs) was 3 178 in sediment and 526 in air. We used morphological features and phylogenetic analyses of actin (actA), calmodulin (cmdA), internal transcribed spacer regions and intervening 5.8S rRNA (ITS), large subunit (LSU) rDNA, RNA polymerase II second largest subunit (rpb2), translation elongation factor 1-alpha (tef1), and β-tubulin (tub2) genes to identify these isolates. Forty-one species belonging to 17 genera of Ascomycota and two of Basidiomycota were identified, and the genus Aspergillus was most commonly observed in the cave (13 taxa). Twenty-four species were found in sediment (16 exclusives) and 25 species were found in air (17 exclusives). In this study, we introduced a new genus (Pseudolecanicillium gen. nov.) in the family Cordycipitaceae and six new species (14 % of the total taxa identified) of fungal isolates obtained from sediment and air: Aspergillus lebretii sp. nov., Malbranchea cavernosa sp. nov., Pseudohumicola cecavii sp. nov., Pseudolecanicillium caatingaense sp. nov., Talaromyces cavernicola sp. nov., and Tritirachium brasiliense sp. nov. In addition, we built a checklist of the fungal taxa reported from Brazilian caves. Our results highlight the contribution of Brazilian caves to the estimation of national and global fungal diversity. Citation: Alves VCS, Lira RA, Lima JMS, Barbosa RN, Bento DM, Barbier E, Bernard E, Souza-Motta CM, Bezerra JDP (2022). Unravelling the fungal darkness in a tropical cave: richness and the description of one new genus and six new species. Fungal Systematics and Evolution 10: 139-167. doi: 10.3114/fuse.2022.10.06.
Keywords: Aspergillus; Caatinga dry forest; cave environment; fungal taxonomy; novel taxa.
© 2022 Westerdijk Fungal Biodiversity Institute.
Conflict of interest statement
Conflict of interest: The authors declare that there is no conflict of interest.
Figures










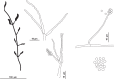
Similar articles
-
Aspergillus, Penicillium, and Talaromyces (Eurotiales) in Brazilian caves, with the description of four new species.Fungal Syst Evol. 2024 Dec;14:89-107. doi: 10.3114/fuse.2024.14.06. Epub 2024 Mar 15. Fungal Syst Evol. 2024. PMID: 39830302 Free PMC article.
-
Living in the dark: Bat caves as hotspots of fungal diversity.PLoS One. 2020 Dec 4;15(12):e0243494. doi: 10.1371/journal.pone.0243494. eCollection 2020. PLoS One. 2020. PMID: 33275627 Free PMC article.
-
Ticket to ride: fungi from bat ectoparasites in a tropical cave and the description of two new species.Braz J Microbiol. 2022 Dec;53(4):2077-2091. doi: 10.1007/s42770-022-00841-y. Epub 2022 Oct 20. Braz J Microbiol. 2022. PMID: 36264483 Free PMC article.
-
Endophytic Diaporthe species from Brazil.Fungal Syst Evol. 2024 Dec;14:251-269. doi: 10.3114/fuse.2024.14.16. Epub 2024 Aug 2. Fungal Syst Evol. 2024. PMID: 39830304 Free PMC article.
-
Unveiling the Subterranean Symphony: A Comprehensive Study of Cave Fungi Revealed Through National Center for Biotechnology Sequences.J Fungi (Basel). 2025 Apr 5;11(4):286. doi: 10.3390/jof11040286. J Fungi (Basel). 2025. PMID: 40278107 Free PMC article. Review.
Cited by
-
New treasures in Cordycipitaceae: Fungicolous fungi associated with Pseudocercospora fijiensis and P. musae in Brazil, including Matutinistella gen. nov.Fungal Syst Evol. 2025 Jun;15:133-152. doi: 10.3114/fuse.2025.15.06. Epub 2024 Oct 18. Fungal Syst Evol. 2025. PMID: 40170761 Free PMC article.
-
Assessing the threat of bat-associated fungal pathogens.One Health. 2023 Apr 28;16:100553. doi: 10.1016/j.onehlt.2023.100553. eCollection 2023 Jun. One Health. 2023. PMID: 37363244 Free PMC article. Review.
-
Three new species of Talaromyces sect. Talaromyces discovered in China.PeerJ. 2024 Oct 11;12:e18253. doi: 10.7717/peerj.18253. eCollection 2024. PeerJ. 2024. PMID: 39403189 Free PMC article.
-
Deconstructing the Dimensions of Mycobiome Fingerprints in Luohandu Cave, Guilin, Southern China.Microorganisms. 2024 Jan 20;12(1):211. doi: 10.3390/microorganisms12010211. Microorganisms. 2024. PMID: 38276196 Free PMC article.
-
Soil ascomycetes from Spain. XIV. The Chaetomiaceae of La Palma (Canary Islands).Persoonia. 2025 Jun;54:93-117. doi: 10.3114/persoonia.2025.54.03. Epub 2025 May 14. Persoonia. 2025. PMID: 40746711 Free PMC article.
References
-
- Auler A, Zogbi L. (2005). Espeleologia: noções básicas. 1st edn. Redespeleo Brasil, Brasil.
-
- Barbosa RN, Bezerra JDP, Costa PMO, et al. (2016). Aspergillus and Penicillium (Eurotiales: Trichocomaceae) in soils of the Brazilian tropical dry forest: diversity in an area of environmental preservation. Revista de Biología Tropical 64: 45–53. - PubMed
-
- Barbosa RN, Bezerra JDP, Santos ACS, et al. (2020). Brazilian tropical dry forest (Caatinga) in the spotlight: an overview of species of Aspergillus, Penicillium and Talaromyces (Eurotiales) and the description of P. vascosobrinhous sp. nov. Acta Botanica Brasilica 34: 409–429.
-
- Bastian F, Jurado V, Nováková A, et al. (2010). The microbiology of Lascaux Cave. Microbiology 3: 644–652. - PubMed
-
- Bento DM, Cruz JB, Santos DJ, et al. (2013). Parque Nacional da Furna Feia – o parque nacional com a maior quantidade de cavernas do Brasil. 32th Congresso Brasileiro de Espeleologia, Barreiras-BA. SBE Campinas: 31–43.
LinkOut - more resources
Full Text Sources
