Enhancer RNA-based modeling of adverse events and objective responses of cancer immunotherapy reveals associated key enhancers and target genes
- PMID: 36741695
- PMCID: PMC9893284
- DOI: 10.3389/fonc.2022.1048127
Enhancer RNA-based modeling of adverse events and objective responses of cancer immunotherapy reveals associated key enhancers and target genes
Abstract
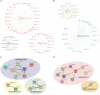
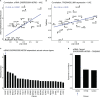
Immune checkpoint inhibitors (ICI) targeting PD-1/PD-L1 or CTLA-4 are emerging and effective immunotherapy strategies. However, ICI-treated patients present heterogeneous responses and adverse events, thus demanding effective ways to assess benefit over risk before treatment. Here, by integrating pan-cancer clinical and molecular data, we tried to predict immune-related adverse events (irAEs, risk) and objective response rates (ORRs, benefit) based on enhancer RNAs (eRNAs) expression among patients receiving anti-PD-1/PD-L1 therapies. We built two tri-variate (eRNAs) regression models, one (with ENSR00000326714, ENSR00000148786, and ENSR00000005553) explaining 71% variance (R=0.84) of irAEs and the other (with ENSR00000164478, ENSR00000035913, and ENSR00000167231) explaining 79% (R=0.89) of ORRs. Interestingly, target genes of irAE-related enhancers, including upstream regulators of MYC, were involved in metabolism, inflammation, and immune activation, while ORR-related enhancers target PAK2 and DLG1 which participate in T cell activation. More importantly, we found that ENSR00000148786 probably enhanced TMEM43/LUMA expression mainly in B cells to induce irAEs in ICI-treated patients. Our study provides references for the identification of immunotherapy-related biomarkers and potential therapeutic targets during immunotherapy.
Keywords: TCGA; TMEM43/LUMA; adverse effect; drug responses; enhancer RNA (eRNA); immune checkpoint block therapy; pan-cancer analysis.
Copyright © 2023 Guo, Lu and Xiong.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

