PLATERO: A calibration protocol for plate reader green fluorescence measurements
- PMID: 36741754
- PMCID: PMC9895789
- DOI: 10.3389/fbioe.2023.1104445
PLATERO: A calibration protocol for plate reader green fluorescence measurements
Abstract
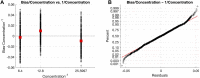
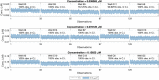
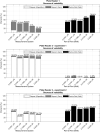
One of the most common sources of information in Synthetic Biology is the data coming from plate reader fluorescence measurements. These experiments provide a measure of the light emitted by a certain fluorescent molecule, such as the Green Fluorescent Protein (GFP). However, these measurements are generally expressed in arbitrary units and are affected by the measurement device gain. This limits the range of measurements in a single experiment and hampers the comparison of results among experiments. In this work, we describe PLATERO, a calibration protocol to express fluorescence measures in concentration units of a reference fluorophore. The protocol removes the gain effect of the measurement device on the acquired data. In addition, the fluorescence intensity values are transformed into units of concentration using a Fluorescein calibration model. Both steps are expressed in a single mathematical expression that returns normalized, gain-independent, and comparable data, even if the acquisition was done at different device gain levels. Most important, the PLATERO embeds a Linearity and Bias Analysis that provides an assessment of the uncertainty of the model estimations, and a Reproducibility and Repeatability analysis that evaluates the sources of variability originating from the measurements and the equipment. All the functions used to build the model, exploit it with new data, and perform the uncertainty and variability assessment are available in an open access repository.
Keywords: fluorescence measurements; measurement data standardization; measurement system analysis; plate reader; units conversion model.
Copyright © 2023 González-Cebrián, Borràs-Ferrís, Boada, Vignoni, Ferrer and Picó.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




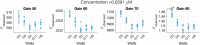




References
LinkOut - more resources
Full Text Sources
Other Literature Sources

