Molecular dynamics simulation of an entire cell
- PMID: 36742032
- PMCID: PMC9889929
- DOI: 10.3389/fchem.2023.1106495
Molecular dynamics simulation of an entire cell
Abstract
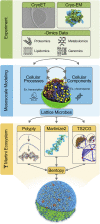
The ultimate microscope, directed at a cell, would reveal the dynamics of all the cell's components with atomic resolution. In contrast to their real-world counterparts, computational microscopes are currently on the brink of meeting this challenge. In this perspective, we show how an integrative approach can be employed to model an entire cell, the minimal cell, JCVI-syn3A, at full complexity. This step opens the way to interrogate the cell's spatio-temporal evolution with molecular dynamics simulations, an approach that can be extended to other cell types in the near future.
Keywords: JCVI-syn3A; Martini force field; coarse grain; integrative modeling; minimal cell; polyply.
Copyright © 2023 Stevens, Grünewald, van Tilburg, König, Gilbert, Brier, Thornburg, Luthey-Schulten and Marrink.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Abraham M. J., Murtola T., Schulz R., Pall S., Smith J. C., Hess B., et al. (2015). ‘GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers’. SoftwareX 1–2, 19–25. 10.1016/j.softx.2015.06.001 - DOI
-
- Alessandri R., Barnoud J., Gertsen A. S., Patmanidis I., de Vries A. H., Souza P. C. T., et al. (2022). Martini 3 coarse-grained force field: Small molecules. Adv. Theory Simulations 5 (1), 2100391. 10.1002/adts.202100391 - DOI
-
- Bhat N. G., Balaji S. (2020). Whole-cell modeling and simulation: A brief survey. New Gener. Comput. 38 (1), 259–281. 10.1007/s00354-019-00066-y - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

