New methods for the quantification of mixed chimerism in transplantation
- PMID: 36742303
- PMCID: PMC9892455
- DOI: 10.3389/fimmu.2023.1023116
New methods for the quantification of mixed chimerism in transplantation
Abstract
Background: Quantification of chimerism showing the proportion of the donor in a recipient is essential for the follow-up of hematopoietic stem cell transplantation but can also be useful to document an immune tolerance situation after solid organ transplantation. Historically, chimerism has been quantified from genomic DNA, but with technological advances, chimerism from donor-derived cell-free DNA seems particularly relevant in solid organ transplantation.
Methods: The reference method was until recently the short tandem repeat technique, but new innovative techniques as digital PCR (dPCR) and NGS, have revolutionized the quantification of chimerism, such as the so-called microchimerism analysis. After a short review of chimerism methods, a comparison of chimerism quantification data for two new digital PCR systems (QIAcuity™ dPCR (Qiagen®) and QuantStudio Absolute Q (ThermoFisher®) and two NGS-based chimerism quantification methods (AlloSeq HCT™ (CareDx®) and NGStrack™ (GenDX®)) was performed.
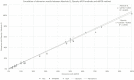
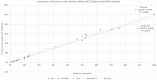
Results: These new methods were correlated and concordant to routinely methods (r²=0.9978 and r²=0.9974 for dPCR methods, r²=0.9978 and r²=0.9988 for NGS methods), and had similar high performance (sensitivity, reproductibility, linearity).
Conclusion: Finally, the choice of the innovative method of chimerism within the laboratory does not depend on the analytical performances because they are similar but mainly on the amount of activity and the access to instruments and computer services.
Keywords: NGS (next generation sequencing); cfDNA (cell-free DNA); chimerism; dPCR (digital PCR); qPCR (quantitative PCR).
Copyright © 2023 Picard, Frassati, Cherouat, Maioli, Moskovtchenko, Cherel, Chiaroni and Pedini.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Yam PY, Petz LD, Knowlton RG, Wallace RB, Stock AD, de Lange G, et al. . Use of DNA restriction fragment length polymorphisms to document marrow engraftment and mixed hematopoietic chimerism following bone marrow transplantation. Transplantation (1987) 43(3):399–407. doi: 10.1097/00007890-198703000-00016 - DOI - PubMed
-
- Casarino L, Carbone C, Capucci MA, Izzi T, Ferrara GB. Analysis of chimerism after bone marrow transplantation using specific oligonucleotide probes. Bone Marrow Transplant (1992) 10(2):165–70. - PubMed
-
- Leclair B, Frégeau CJ, Aye MT, Fourney RM. DNA Typing for bone marrow engraftment follow-up after allogeneic transplant: A comparative study of current technologies. Bone Marrow Transplant (1995) 16(1):43–55. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

