Transcriptome analysis provides StMYBA1 gene that regulates potato anthocyanin biosynthesis by activating structural genes
- PMID: 36743487
- PMCID: PMC9895859
- DOI: 10.3389/fpls.2023.1087121
Transcriptome analysis provides StMYBA1 gene that regulates potato anthocyanin biosynthesis by activating structural genes
Abstract
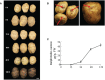
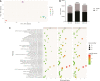
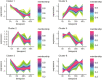
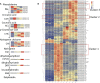
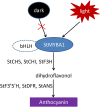
Anthocyanin biosynthesis is affected by light, temperature, and other environmental factors. The regulation mode of light on anthocyanin synthesis in apple, pear, tomato and other species has been reported, while not clear in potato. In this study, potato RM-210 tubers whose peel will turn purple gradually after exposure to light were selected. Transcriptome analysis was performed on RM-210 tubers during anthocyanin accumulation. The expression of StMYBA1 gene continued to increase during the anthocyanin accumulation in RM-210 tubers. Moreover, co-expression cluster analysis of differentially expressed genes showed that the expression patterns of StMYBA1 gene were highly correlated with structural genes CHS and CHI. The promoter activity of StMYBA1 was significantly higher in light conditions, and StMYBA1 could activate the promoter activity of structural genes StCHS, StCHI, and StF3H. Further gene function analysis found that overexpression of StMYBA1 gene could promote anthocyanin accumulation and structural gene expression in potato leaves. These results demonstrated that StMYBA1 gene promoted potato anthocyanin biosynthesis by activating the expression of structural genes under light conditions. These findings provide a theoretical basis and genetic resources for the regulatory mechanism of potato anthocyanin synthesis.
Keywords: StMYBA1; anthocyanin biosynthesis; light-induced; potato; transcriptional activation.
Copyright © 2023 Zhao, Zhang, Liu, Zhao, Hu, Liu, Lin, Song and He.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
LinkOut - more resources
Full Text Sources

