DOF transcription factors: Specific regulators of plant biological processes
- PMID: 36743498
- PMCID: PMC9897228
- DOI: 10.3389/fpls.2023.1044918
DOF transcription factors: Specific regulators of plant biological processes
Abstract
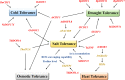
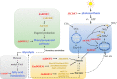
Plant biological processes, such as growth and metabolism, hormone signal transduction, and stress responses, are affected by gene transcriptional regulation. As gene expression regulators, transcription factors activate or inhibit target gene transcription by directly binding to downstream promoter elements. DOF (DNA binding with One Finger) is a classic transcription factor family exclusive to plants that is characterized by its single zinc finger structure. With breakthroughs in taxonomic studies of different species in recent years, many DOF members have been reported to play vital roles throughout the plant life cycle. They are not only involved in regulating hormone signals and various biotic or abiotic stress responses but are also reported to regulate many plant biological processes, such as dormancy, tissue differentiation, carbon and nitrogen assimilation, and carbohydrate metabolism. Nevertheless, some outstanding issues remain. This article mainly reviews the origin and evolution, protein structure, and functions of DOF members reported in studies published in many fields to clarify the direction for future research on DOF transcription factors.
Keywords: DOF; metabolic regulation; plant hormones; transcription factor; zinc finger.
Copyright © 2023 Zou and Sun.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Boccaccini A., Lorrai R., Ruta V., Frey A., Mercey-Boutet S., Marion-Poll A., et al. (2016). The DAG1 transcription factor negatively regulates the seed-to-seedling transition in Arabidopsis acting on ABA and GA levels. BMC Plant Biol. 16 (1), 198. doi: 10.1186/s12870-016-0890-5 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Research Materials

