Functional genomic analysis of K+ related salt-responsive transporters in tolerant and sensitive genotypes of rice
- PMID: 36743539
- PMCID: PMC9893783
- DOI: 10.3389/fpls.2022.1089109
Functional genomic analysis of K+ related salt-responsive transporters in tolerant and sensitive genotypes of rice
Abstract
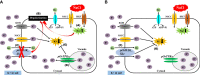
Introduction: Salinity is a complex environmental stress that affects the growth and production of rice worldwide. But there are some rice landraces in coastal regions that can survive in presence of highly saline conditions. An understanding of the molecular attributes contributing to the salinity tolerance of these genotypes is important for developing salt-tolerant high yielding modern genotypes to ensure food security. Therefore, we investigated the role and functional differences of two K+ salt-responsive transporters. These are OsTPKa or Vacuolar two-pore potassium channel and OsHAK_like or a hypothetical protein of the HAK family. These transporters were selected from previously identified QTLs from the tolerant rice landrace genotype (Horkuch) and sensitive genotype (IR29).
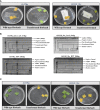
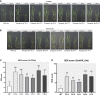
Methods: In silico comparative sequence analysis of the promoter sequences of two these genes between Horkuch and IR29 was done. Real-Time expression of the selected genes in leaves and roots of IR29 (salt-sensitive), I-14 and I-71 (Recombinant Inbred Lines of IR29(♀)× Horkuch), Horkuch and Pokkali (salt-tolerant) under salt-stress at different time points was analyzed. For further insight, OsTPKa and OsHAK_like were chosen for loss-of-function genomic analysis in Horkuch using the CRISPR/Cas9 tool. Furthermore, OsTPKa was chosen for cloning into a sensitive variety by Gateway technology to observe the effect of gain-of-function.
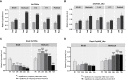
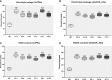
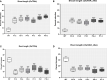
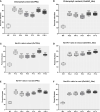
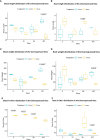
Results: The promoter sequences of the OsTPKa and OsHAK_like genes showed some significant differences in promoter sequences which may give a survival advantage to Horkuch under salt-stress. These two genes were also found to be overexpressed in tolerant varieties (Horkuch and Pokkali). Moreover, a coordinated expression pattern between these two genes was observed in tolerant Horkuch under salt-stress. Independently transformed plants where the expression of these genes was significantly lowered, performed poorly in physiological tests for salinity tolerance. On the other hand, positively transformed T0 plants with the OsTPKa gene from Horkuch consistently showed growth advantage under both control and salt stress.
Discussion: The poor performance of the transgenic plants with the down-regulated genes OsTPKa and OsHAK_like under salt stress supports the assumption that OsTPKa and OsHAK_like play important roles in defending the rice landrace Horkuch against salt stress, minimizing salt injury, and maintaining plant growth. Moreover, the growth advantage provided by overexpression of the vacuolar OsTPKa K+ transporter, particularly under salt stress reconfirms its important role in providing salt tolerance. The QTL locus from Horkuch containing these two transporters maybe bred into commercial rice to produce high-yielding salt tolerant rice.
Keywords: CRISPR/Cas9; Horkuch; OsHAK_like; OsTPKa; expression analysis; rice; salt-sensitive; salt-tolerant.
Copyright © 2023 Haque, Elias, Jahan and Seraj.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Ahmadizadeh M., Vispo N. A., Calapit-Palao C. D. O., Pangaan I. D., Viña C. D., Singh R. K. (2016). Reproductive stage salinity tolerance in rice: a complex trait to phenotype. Indian J. Plant Physiol. 21, 528–536. doi: 10.1007/s40502-016-0268-6 - DOI
-
- Ahmed T., Biswas S., Elias S. M., Rahman M. S., Tuteja N., Seraj Z. I. (2018). In planta transformation for conferring salt tolerance to a tissue-culture unresponsive indica rice (Oryza sativa l.) cultivar. In Vitro Cell.Dev.Biol.-Plant 54, 154–165. doi: 10.1007/s11627-017-9870-1 - DOI
-
- Amin M., Elias S. M., Hossain A., Ferdousi A., Rahman M. S., Tuteja N., et al. (2012). Over-expression of a DEAD-box helicase, PDH45, confers both seedling and reproductive stage salinity tolerance to rice (Oryza sativa l.). Mol. Breed. 30, 345–354. doi: 10.1007/s11032-011-9625-3 - DOI
-
- Amirjani M. R. (2011). Effect of salinity stress on growth, sugar content, pigments and enzyme activity of rice. Int. J. Bot. 7, 73–81. doi: 10.3923/ijb.2011.73.81 - DOI
LinkOut - more resources
Full Text Sources
Research Materials

